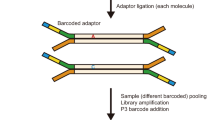
Abstract
We have developed a protocol for the generation of genome-wide maps (meiomaps) of recombination and chromosome segregation for the three products of human female meiosis: the first and second polar bodies (PB1 and PB2) and the corresponding oocyte. PB1 is biopsied and the oocyte is artificially activated by exposure to calcium ionophore, after which PB2 is biopsied and collected with the corresponding oocyte. The whole genomes of the polar bodies and oocytes are amplified by multiple displacement amplification and, together with maternal genomic DNA, genotyped for ∼300,000 single-nucleotide polymorphisms (SNPs) genome-wide by microarray. Informative maternal heterozygous SNPs are phased using a haploid PB2 or oocyte as a reference. A simple algorithm is then used to identify the maternal haplotypes for each chromosome, in all of the products of meiosis for each oocyte. This allows mapping of crossovers and analysis of chromosome segregation patterns. The protocol takes a minimum of 3–5 d and requires a clinical embryologist with micromanipulation experience and a molecular biologist with basic bioinformatic skills. It has several advantages over previous methods; importantly, the use of artificial oocyte activation avoids the creation of embryos for research purposes. In addition, compared with next-generation sequencing, targeted SNP genotyping is cost-effective and it simplifies the bioinformatic analysis, as only one haploid reference sample is required to establish phase for maternal haplotyping. Finally, meiomapping is more informative than copy-number analysis alone for analysis of chromosome segregation patterns. Using this protocol, we have provided new insights that may lead to improvements in assisted reproduction for the treatment of infertility.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hassold, T. & Hunt, P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 (2001).
Nagaoka, S.I., Hassold, T.J. & Hunt, P.A. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13, 493–504 (2012).
Angell, R. First-meiotic-division nondisjunction in human oocytes. Am. J. Hum. Genet. 61, 23–32 (1997).
Handyside, A.H. et al. Multiple meiotic errors caused by predivision of chromatids in women of advanced maternal age undergoing in vitro fertilisation. Eur. J. Hum. Genet. 20, 742–747 (2012).
Handyside, A.H. et al. Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J. Med. Genet. 47, 651–658 (2010).
Natesan, S.A. et al. Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet. Med. 16, 838–845 (2014).
Ottolini, C.S. et al. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat. Genet. 47, 727–735 (2015).
Hou, Y. et al. Genome analyses of single human oocytes. Cell 155, 1492–1506 (2013).
Fulton, B.P. & Whittingham, D.G. Activation of mammalian oocytes by intracellular injection of calcium. Nature 273, 149–151 (1978).
Winston, N., Johnson, M., Pickering, S. & Braude, P. Parthenogenetic activation and development of fresh and aged human oocytes. Fertil. Steril. 56, 904–912 (1991).
Taylor, A.S. & Braude, P.R. The early development and DNA content of activated human oocytes and parthenogenetic human embryos. Hum. Reprod. 9, 2389–2397 (1994).
Liu, Y. et al. Three-day-old human unfertilized oocytes after in vitro fertilization/intracytoplasmic sperm injection can be activated by calcium ionophore a23187 or strontium chloride and develop to blastocysts. Cell. Reprogram. 16, 276–280 (2014).
de Fried, E.P. et al. Human parthenogenetic blastocysts derived from noninseminated cryopreserved human oocytes. Fertil. Steril. 89, 943–947 (2008).
Escrich, L. et al. Spontaneous in vitro maturation and artificial activation of human germinal vesicle oocytes recovered from stimulated cycles. J. Assist. Reprod. Genet. 28, 111–117 (2011).
Montag, M., Koster, M., van der Ven, K., Bohlen, U. & van der Ven, H. The benefit of artificial oocyte activation is dependent on the fertilization rate in a previous treatment cycle. Reprod. Biomed. Online 24, 521–526 (2012).
Kuentz, P. et al. Assisted oocyte activation overcomes fertilization failure in globozoospermic patients regardless of the DPY19L2 status. Hum. Reprod. 28, 1054–1061 (2013).
Heindryckx, B., Van der Elst, J., De Sutter, P. & Dhont, M. Treatment option for sperm- or oocyte-related fertilization failure: assisted oocyte activation following diagnostic heterologous ICSI. Hum. Reprod. 20, 2237–2241 (2005).
Heindryckx, B., De Gheselle, S., Gerris, J., Dhont, M. & De Sutter, P. Efficiency of assisted oocyte activation as a solution for failed intracytoplasmic sperm injection. Reprod. Biomed. Online 17, 662–668 (2008).
Ebner, T. et al. Treatment with Ca2+ ionophore improves embryo development and outcome in cases with previous developmental problems: a prospective multicenter study. Hum. Reprod. 30, 97–102 (2015).
Hassold, T., Merrill, M., Adkins, K., Freeman, S. & Sherman, S. Recombination and maternal age-dependent nondisjunction: molecular studies of trisomy 16. Am. J. Hum. Genet. 57, 867–874 (1995).
Oliver, T.R. et al. New insights into human nondisjunction of chromosome 21 in oocytes. PLoS Genet. 4, e1000033 (2008).
Middlebrooks, C.D. et al. Evidence for dysregulation of genome-wide recombination in oocytes with nondisjoined chromosomes 21. Hum. Mol. Genet. 2, 408–417 (2013).
Kong, A. et al. Recombination rate and reproductive success in humans. Nat. Genet. 36, 1203–1206 (2004).
Gruhn, J.R., Rubio, C., Broman, K.W., Hunt, P.A. & Hassold, T. Cytological studies of human meiosis: sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS One 8, e85075 (2013).
Tease, C., Hartshorne, G.M. & Hulten, M.A. Patterns of meiotic recombination in human fetal oocytes. Am. J. Hum. Genet. 70, 1469–1479 (2002).
Holloway, J.K., Booth, J., Edelmann, W., McGowan, C.H. & Cohen, P.E. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4, e1000186 (2008).
Zaragoza, M.V. et al. Nondisjunction of human acrocentric chromosomes: studies of 432 trisomic fetuses and liveborns. Hum. Genet. 94, 411–417 (1994).
Capalbo, A. et al. Sequential comprehensive chromosome analysis on polar bodies, blastomeres and trophoblast: insights into female meiotic errors and chromosomal segregation in the preimplantation window of embryo development. Hum. Reprod. 28, 509–518 (2013).
Christopikou, D. et al. Polar body analysis by array comparative genomic hybridization accurately predicts aneuploidies of maternal meiotic origin in cleavage stage embryos of women of advanced maternal age. Hum. Reprod. 28, 1426–1434 (2013).
Palermo, G.D., Cohen, J., Alikani, M., Adler, A. & Rosenwaks, Z. Development and implementation of intracytoplasmic sperm injection (ICSI). Reprod. Fertil. Dev. 7, 211–217 (1995).
Findlay, I., Ray, P., Quirke, P., Rutherford, A. & Lilford, R. Allelic drop-out and preferential amplification in single cells and human blastomeres: implications for preimplantation diagnosis of sex and cystic fibrosis. Hum. Reprod. 10, 1609–1618 (1995).
Bonetti, A. et al. Ultrastructural evaluation of human metaphase II oocytes after vitrification: closed versus open devices. Fertil. Steril. 95, 928–935 (2011).
Kuwayama, M. Highly efficient vitrification for cryopreservation of human oocytes and embryos: the Cryotop method. Theriogenology 67, 73–80 (2007).
Alexander, M. & Kusleika, D. Excel 2016 Power Programming with VBA. (Chichester, UK: John Wiley & Sons, 2016).
Author information
Authors and Affiliations
Contributions
C.S.O., A.C., D.C., L.R. and F.M.U. were responsible for donor consent, oocyte collection and oocyte activation. L.R., F.M.U. and A.H.H. oversaw ethical and legal regulations in Italy and the UK. A.C., C.S.O., S.A.N. and D.C. carried out amplification, SNP array and array CGH experiments. A.H.H., L.N. and C.S.O. analyzed the SNP data. A.H.H., C.S.O. and L.N. generated the figures. A.H.H., L.N. and C.S.O. wrote the manuscript. A.H.H., E.R.H. and L.N. edited the manuscript. All authors proofread and accepted the manuscript. A.H.H. developed the meiomapping algorithm.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Data
Meiomap SNP data (XLSM 1464 kb)
Rights and permissions
About this article
Cite this article
Ottolini, C., Capalbo, A., Newnham, L. et al. Generation of meiomaps of genome-wide recombination and chromosome segregation in human oocytes. Nat Protoc 11, 1229–1243 (2016). https://doi.org/10.1038/nprot.2016.075
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.075
This article is cited by
-
Genetically-biased fertilization in APOBEC1 complementation factor (A1cf) mutant mice
Scientific Reports (2022)
-
Molecular basis of reproductive senescence: insights from model organisms
Journal of Assisted Reproduction and Genetics (2021)
-
Karyomapping in preimplantation genetic testing for β-thalassemia combined with HLA matching: a systematic summary
Journal of Assisted Reproduction and Genetics (2019)
-
Tripolar mitosis and partitioning of the genome arrests human preimplantation development in vitro
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.