Abstract
Genetically encoded calcium indicators for visualizing dynamic cellular activity have greatly expanded our understanding of the brain. However, owing to the light-scattering properties of the brain, as well as the size and rigidity of traditional imaging technology, in vivo calcium imaging has been limited to superficial brain structures during head-fixed behavioral tasks. These limitations can now be circumvented by using miniature, integrated microscopes in conjunction with an implantable microendoscopic lens to guide light into and out of the brain, thus permitting optical access to deep brain (or superficial) neural ensembles during naturalistic behaviors. Here we describe steps to conduct such imaging studies using mice. However, we anticipate that the protocol can be easily adapted for use in other small vertebrates. Successful completion of this protocol will permit cellular imaging of neuronal activity and the generation of data sets with sufficient statistical power to correlate neural activity with stimulus presentation, physiological state and other aspects of complex behavioral tasks. This protocol takes 6–11 weeks to complete.
Similar content being viewed by others
Introduction
Methods for monitoring neural activity in vivo, such as electrophysiology1,2, neurochemical measurements3,4 and optical imaging of fluorescently reported neural dynamics5,6,7,8, have substantially increased our understanding of the neural computations underlying complex behavioral states. Among these techniques, the development and implementation of genetically encoded Ca2+ indicators (GECIs)6,9,10,11,12,13 has been particularly advantageous to the study of brain function. During periods of heightened neural activity14,15, Ca2+ flows into the dendritic branches16 and cell bodies of neurons, increasing intracellular concentrations of Ca2+. These activity-dependent fluctuations in intracellular Ca2+ can be monitored by introducing a Ca2+ indicator, such as GCaMP13, into the neuronal population of interest. Upon the binding of Ca2+, indicators such as GCaMP emit a fluorescent signal, which is intensified during periods of elevated neural activity14,15. As a result, fluctuations in fluorescence emission over time can be used to infer dynamic changes in cellular activity. Moreover, when these signals are visualized with cellular-level resolution, fluorescent indicators of neural activity provide not only temporal information related to dynamic changes in brain cell activity17, but they also enable the visualization of each cell's spatial location within the brain18.
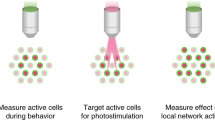
The visualization of fluorescently reported neural signals within the brain requires both a light source to excite the Ca2+ indicator and a light-sensing device to detect the emission signal19. As a result, optical monitoring of fluorescently reported neural activity is constrained by physical limitations to transmit light to and from the neuron population of interest. Specifically, neural tissue exhibits high levels of light scattering, and this physical property has precluded optical imaging of in vivo Ca2+ transients within deep brain regions19. However, limitations in the propensity to transmit light through the brain can be circumvented by the implantation of a microendoscopic lens (gradient-refractive-index (GRIN) lenses attached on either end of a relay lens) in subcortical or deep brain regions20,21,22 (Fig. 1a). Microendoscopic lenses facilitate the visualization of deep-brain neural activity by relaying light to and from deep brain structures, bringing previously inaccessible fluorescence signals into a field of view outside the brain11,23,24,25. For example, these lenses can be used to visualize the activity of genetically defined neurons located within deep brain regions, such as the hypothalamus, thus greatly expanding the population of neurons that can be visualized and monitored within the brain26,27.
(a) Cartoon diagram of a miniature, integrated microscope containing all optical components necessary for the visualization of fluorescently encoded Ca2+ transients in freely behaving mice. Specifically, the microscope contains an excitation light source (473-nm LED), lenses for guiding and focusing light, and a fluorescence sensor to detect emission light; it is small enough to fit on the head of a mouse (inset shows the actual size of the microscope). To visualize GCaMP-expressing neuron activity within deep brain regions, this microscope can be used in conjunction with a microendoscope chronically implanted in the brain. (b) Example of digitally captured fluorescence activity from the brain of a freely behaving mouse. The green arrow points to a GCaMP6-expressing neuron and the red arrow points to a clearly visualized blood vessel. Scale bar, 123 μm. (c) Example data extracted from digitally captured fluorescence signals. Image on the left shows the spatial location of identified cells, and traces to the right show changes in fluorescence signal over time, represented as the variance from the mean fluorescence signal at each time point. Zoomed-in portion of traces shows example Ca2+ activity traces from a cell that increased firing (top, green) and an example of a cell that decreased firing (bottom, yellow) in response to the presentation of a social stimulus. Representative data were collected from one mouse in one acute recording session. All procedures were approved by the UNC IACUC.
Owing to the size and rigidity of traditional imaging technology, previous imaging studies have also been limited in the types of behaviors that could be examined. However, the development of miniature, integrated microscopes, which are small and light enough to be carried on the head of the animal without interfering with behavior, has greatly expanded the scope of scientific questions that can be asked28. Importantly, these miniature microscopes can be used in conjunction with microendoscopic lenses, permitting access to deep brain neural computations associated with naturalistic animal behavior (Fig. 1a)22,26,27. An additional advantage of this technique is that it can be used to repeatedly visualize large-scale neuronal populations (up to 1,000 neurons in one mouse29), thus providing a substantial increase in statistical power over previously developed in vivo recording methods. Here we provide a detailed protocol for the surgical implantation of a microendoscopic lens above a deep brain (or superficial) neuron population and the subsequent collection of in vivo imaging data in a freely behaving mouse (Fig. 1b,c). These procedures have been developed and used by our laboratory to successfully image neuronal populations during active, naturalistic, animal behaviors26.
Comparison with other in vivo recording methods
One commonly used approach to image fluorescently reported neuronal dynamics is two-photon microscopy30. This technique uses low-energy, near-infrared (IR) photons to penetrate highly light-scattering brain tissue up to 600–700 μm below the surface of the brain31. A major advantage of two-photon microscopy is its ability to selectively excite fluorophores within a well-defined focal plane, which results in a spatial resolution capable of resolving cellular activity within precisely defined anatomical subregions of neurons, such as dendrites and axonal boutons30. Notably, although this imaging modality provides superior spatial resolution, it requires the head fixation of animals and, in the absence of a microendoscope or optical cannula, two-photon imaging is limited to superficial layers of the brain32,33. Together, these behavioral and optical limitations greatly reduce the scope of scientific questions that can be examined with two-photon microscopy.
Implantation of small, lightweight fiber optics above a region of interest, such as with fiber photometry, circumvents optical and behavioral limitations posed by two-photon microscopy34. However, unlike two-photon microscopy, fiber photometry lacks cellular-level resolution, and it provides only aggregate activity within the field of view (i.e., bulk changes in fluorescent signal)22. Thus, this method is better suited for monitoring dynamic activity within neural projection fields35. In addition to limitations in optical resolution, fiber photometry requires the test subject to be secured to a rigid fiber optic bundle, which can be difficult for small mammals, such as mice, to maneuver34. Thus, although fiber photometry increases the depth in which neural activity can be monitored, it presents substantial limitations in optical resolution, restricts the natural behavioral repertoire of an animal and limits the animal models that can be optimally used.
Large-scale recordings of neural activity within freely behaving mammals36 can also be conducted with techniques that do not rely on the use of fluorescence indicators of neural activity, such as in vivo electrophysiological recordings2. Importantly, compared with in vivo Ca2+ imaging, electrophysiology provides superior temporal resolution, allowing for more-accurate spike timing estimations17,37,38, as well as the correlation of neural activity with precisely defined temporal events. In addition, in vivo electrophysiology can be combined with optogenetic perturbations of genetically defined neuronal populations to permit the identification (although not unequivocally) and manipulation of defined neuronal populations39,40,41. The ability to monitor and subsequently manipulate a circuit is particularly important to the study of brain function, as it allows the causal role of identified computations to be elucidated. Thus, compared with freely behaving in vivo optical imaging methods, in vivo electrophysiology methods offer advantages in the domain of temporal resolution, as well as technological integration. One notable limitation of this method is that the spatial location of monitored cells cannot be visualized, which makes it difficult to assert that an identified cell is similar or unique across recording sessions1. Moreover, because in vivo electrophysiology relies on waveform shapes to differentiate individual cells from each other, it can be challenging to detect cells with sparse firing patterns or those that are located within densely populated networks. Finally, the number of cells that can be detected with in vivo electrophysiology methods is often far less than the number of cells that can be monitored with the optical imaging methods described in this protocol29,42. Taken together, these limitations in cell identification and statistical power pose a major disadvantage for studies that require chronic monitoring of neural activity.
Advantages and application of microendoscopic Ca2+ imaging
In an effort to understand both adaptive and maladaptive states, in vivo methods for monitoring neural activity in animal models have been developed to characterize patterns of brain activity associated with adaptive behavioral states, as well as those resembling core features of human psychiatric illness43. Many of the behavioral paradigms used in these studies require animals to be able to move about the testing apparatus in a naturalistic, unrestricted manner. The lightweight, head-mountable microscopes used in this protocol are well suited for such studies, as they allow neuronal activity to be monitored during relatively unrestricted behavior22,28. Another advantage to the microscopes described in this protocol is that they can easily be attached and detached to the skull of an individual animal, thus allowing the same neuronal populations to be repeatedly imaged across multiple experimental sessions29,44. This particular advantage makes this imaging methodology appealing to studies related to neural development, as well as research examining maladaptive neural plasticity associated with the transition to pathological disease states.
Although many previous imaging studies have been limited to superficial areas of the brain, the implantation of a microendoscopic lens as described in this protocol expands the transmission of neural information by light far beyond superficial cortical regions. Given that the cortex contains only about 5.3% of neurons in the mouse brain45, this protocol substantially increases the neuronal populations in which in vivo Ca2+ imaging methods can be applied to the study of brain function. Finally, when a microendoscopic lens is used in combination with a genetically encoded Ca2+ indicator9,26,34,46, neural activity within brain structures composed of heterogeneous populations of neurons can be decoded at the microcircuit level. Together, these advantages will facilitate the dynamic mapping of large-scale neural circuits and possibly the reconstruction of typical activity patterns in the diseased brain19,43.
Limitations
Although this methodology presents many advantages over other in vivo recording procedures, limitations do exist. For example, although the microscope used in this protocol allows for a wide variety of naturalistic behaviors to be examined22,26,29, currently available microscopes are not waterproof, and they are therefore not suitable for behavioral experiments that require the test subject to be submerged in water, such as the forced swim task22 or Morris water maze47. In addition, these microscopes are not compatible for use with far-red-shifted indicators28 that are better suited for combining in vivo imaging methods with optogenetic methods for circuit perturbation39,48,49. The ability to monitor and manipulate a defined neuronal population within the same subject will require technological advances in the light transmission and detection properties of currently available miniature microscopes. This specific advancement will be crucial for determining the causal role of specific neuronal computations involved in the generation of complex behaviors36. Finally, owing to insufficient camera frame rates, currently available microscopes may not be suitable for imaging neural membrane dynamics via voltage sensors19, which provide superior temporal resolution over Ca2+ indicators. Thus, concurrent advancements in both protein engineering and optical imaging devices are needed if this in vivo imaging method is to be used in conjunction with optogenetic manipulations of neural circuits or with indicators that more accurately monitor the temporal dynamics of cellular activity.
Current in vivo imaging methods in freely behaving mammals only allow for transient cellular activity to be captured and analyzed within a subset of an entire neural circuit. Specifically, the sizes of both the implantable microendoscopic GRIN lens and the microscope baseplate docking system limit the detection of in vivo Ca2+ transients with this imaging method to one brain region per animal. Given that molecularly and anatomically defined neuronal ensembles are merely components of larger neural networks spread across multiple brain nuclei, concurrent imaging of multiple brain nuclei is necessary to understand how dynamic activity across an entire neural circuit contributes to both adaptive and pathological behavioral responses50. Finally, it is also important to consider that the implantation of a microendoscopic lens above a target neuron population may damage or disrupt associated neural circuits that are critical for the appropriate expression of the behavior or physiological condition under investigation.
Similarly to the rapid growth in data acquisition methods previously experienced in the fields of physics and genetics, data storage and analysis methods in neuroscience have not kept up with the exponential increases in the rate of data acquisition. To take full advantage of the information encoded in the relatively large and complex data sets that are generated by in vivo Ca2+ imaging, continued advancements are needed in methods for data storage and retrieval; the rate and efficiency in which temporal and spatial information of individual cellular units can be extracted; and the ability to accurately correlate these data with observed environmental, behavioral or physiological events37,51,52,53. For example, a powerful future application of this imaging method would be the potential to monitor, decode and manipulate neural circuit activity within a freely behaving animal in real time. In addition to the advances in microscope technology described above that are necessary to enable concurrent optical monitoring and manipulation of neural activity, the rate at which the dynamic activity of microcircuits can be extracted and decoded would also need to be substantially increased54. Ultimately, the ability to monitor, manipulate and model the collective activity of neural circuits will be essential to decoding the neural syntax that regulates complex physiological and behavioral states.
Experimental design
Animal model and microendoscope selection. The experimental preparation presented in this protocol is ideal for imaging in small mammals, such as mice, but it can also be adapted to larger animal models, such as rats, as well as to other nontraditional laboratory species, such as songbirds55. However, when working with larger animal models, it is important to consider possible limitations in imaging. For example, a microendoscope that is 8 mm in length can easily reach deep brain areas such as the ventral tegmental area in a mouse (depth of brain region: ∼4.15 mm ventral to bregma), but it would not be long enough to reach this same structure in a rat (depth of brain region: ∼7.3 mm ventral to bregma). In this case, it is possible to have a custom microendoscope of appropriate length fabricated (see MATERIALS section for recommended companies), but the optics of the combined microendoscope and microscope system would need to be tested before proceeding with in vivo experiments. Specifically, the microendoscope would need to be designed in a manner that would permit the adequate transmission of light in the range of the excitation and emission spectra of the Ca2+ indicator (Fig. 1), while limiting the optical distortions that can occur with the use of a GRIN lens56.
Introduction of GCaMP. If a viral indicator of GCaMP is being used, a critical step in this procedure is the optimization of viral expression within the target cell population29. This portion of the procedure can take 3–8 weeks, depending on the goals of the experiment, and it should be conducted before the start of in vivo imaging experiments (Fig. 2). To optimize viral expression of GCaMP, we recommend conducting a dilution protocol and that the protocol be carried out to the longest planned time point of the experiment. For example, for experimental designs in which only one acute imaging session is needed, a dilution study at one experimental time point should be sufficient. This time point should be selected on the basis of optimal viral expression of GCaMP (3–8 weeks), the duration of time required to recover from surgical procedures (2–3 weeks), and the learning and/or habituation procedures directly related to the experimental manipulation.
Before beginning in vivo imaging experiments, first conduct a virus dilution study (Steps 1–5). Next, transduce cells with the appropriate dilution of the viral construct and implant a microendoscope above the target neuron population (Steps 6–47). After sufficient time has been allowed for the virus to express and the tissue to clear under the microendoscope, use the miniature, integrated microscope to check the expression levels of the Ca2+ indicator. If cells can be clearly visualized and if there is an indication of dynamic changes in fluorescence activity, perform baseplate attachment surgery (Steps 48–76). If required for behavioral experiments, habituate mice to procedures for attaching a baseplate and in vivo recordings (Step 77). Once the necessary surgical and habituation procedures have been completed, conduct in vivo imaging studies in freely behaving mice and extract data (Steps 78–102).
Given the potential to repeatedly monitor neural activity within the same test subject, it is more likely that this protocol will be used for prolonged optical monitoring of defined neuronal ensembles. For chronic imaging experiments, it is recommended that the dilution protocol be carried out to the longest planned time point (e.g., 8 weeks after stereotaxic injection of virus; Fig. 2) and that this step be completed before beginning an experimental cohort of animals. A failure to optimize viral expression of GCaMP for the total duration of the study may result in overexpression of the indicator at later time points. Overexpression of GCaMP is deleterious to the experimental procedures, as it can substantially interfere with a cell's ability to buffer Ca2+, resulting in a lack of dynamic Ca2+ signals, nuclear filling of GCaMP and cell death14,19,29. Conversely, inadequate expression of the viral indicator would hinder the ability to visualize fluorescently encoded GCaMP signals or the likelihood of extracting statistically significant Ca2+ signals from the data.
Although the methods presented in this protocol emphasize the use of virally encoded GCaMP indicators, transgenic mouse lines engineered to express fluorescent reporters of neural activity57,58,59,60 can also be used. An advantage to these lines is that they stably express GCaMP throughout development, which may be better suited for chronic imaging experiments that are subject to the risk of overexpression of the viral indicator. An additional advantage to the use of a transgenic line engineered to express GCaMP is that there is greater homogeneity in GCaMP expression across subjects. An additional alternative method for the introduction of GCaMP is in utero electroporation61. This method of viral transduction may be better suited for animal models in which few transgenic lines are available62. Before the start of an in vivo imaging experiment, these alternative methods for the visualization of fluorescently reported neural activity should be tested within the target neuron population of interest, as well as in conjunction with the appropriate optic materials (microendoscope and microscope).
Surgical procedures for lens implantation and baseplate attachment. Once the appropriate microendoscope has been selected and expression levels of the fluorescence indicator have been optimized, in vivo imaging experiments can be conducted for weeks to months after the initial surgery, thus permitting long-term tracking of defined neuronal ensembles29. Subsequent procedures to conduct chronic imaging studies within deep brain regions require the surgical implantation of a microendoscopic lens above the target neuron population44 and the attachment of a baseplate to the mouse's (or other species) head to interface with the miniature, integrated microscope23,28. Each of these surgical procedures takes about 2–3 h to complete (Fig. 2).
Behavioral procedures and data analysis. When designing freely behaving in vivo Ca2+ imaging experiments, it is important to consider a list of experimental precautions and controls that will be required to permit accurate data interpretation. For instance, although some movement of neural tissue is normal and it can be corrected for by the application of a motion-correction algorithm63, optical monitoring of neural activity within deep brain regions of freely behaving animals has a heightened potential for behaviorally mediated motion artifacts to be encoded in the data. Although we have not yet found this to be an issue with the preparation described in this protocol, it is possible that during periods of rapid or rigorous animal movement extra strain may be placed on the animal's skull, causing the position of the lens in relation to the target neuron population to shift between frames. These types of mechanically induced shifts in the imaging plane are of particular concern to the experimental design, as they have the potential to alter both the intensity and spatial location of optically detected signals. Thus, during active behavior, it is possible for motion-induced changes in fluorescence signals to be inappropriately assigned as physiologically relevant neural signals.
To ensure that optically detected fluorescent signals are representative of neural activity, care should be taken to ensure that the test subject can freely move throughout its environment, and that it can perform the behavioral task with minimal restraint. In addition, once the data collection process is complete, optically detected fluorescence signals should be inspected to confirm that they have the appropriate morphology and that changes in fluorescence signals over time have identified characteristics of Ca2+ transients (Fig. 3). Finally, it is also possible to include an experimental group that contains a lens above the target neuron population, but rather than introducing a fluorescent reporter of neural activity a fluorescent marker for cell identification, such as GFP, is introduced into the target neuron population. In this case, dynamic changes in neural activity should only be observed in animals expressing the fluorescent reporter of neural activity and not GFP-expressing animals35. Any observed changes in fluorescence in the GFP group would be the result of movement noise and possibly indicate that the design of the experimental paradigm is not optimal for this methodology or that more care needs to be taken during the experimental preparation to ensure that the head cap is securely affixed to the skull.
(a–d) Signals extracted via automated cell sorting algorithms will vary in data quality. (a–d) In the examples shown here, cellular signals are ordered from highest (a) to lowest (d) quality, and they were extracted from digitally acquired Ca2+ imaging data using PCA/ICA analysis (Mosaic analysis software, Inscopix). This automated cell sorting algorithm deconstructs the data set into statistically independent signals (Independent components) composed of a spatial filter that indicates the number and coordinates of pixels that contribute to an identified signal (left), as well as an activity trace that shows how the intensity of pixels within this defined spatial filter change over time (right). (a) Independent components (ICs) that have the characteristics of dynamic fluorescence emissions from a GCaMP-expressing cell should have a spatial filter with morphological features of a cell body (i.e., a spherical shape), as well as an activity trace that resembles the temporal dynamics of a Ca2+ transient (i.e., a fast rise time and slow offset). This IC also has relatively low levels of background fluorescence compared with the large changes in signal fluctuation that occur during the three Ca2+ events (red stars) shown in the activity trace, indicating a high signal-to-noise ratio. Negative fluctuations in fluorescence emissions can also be seen within this activity trace (gray stars). Although it is tempting to assign negative fluctuations in fluorescence intensity to decreases in neural activity for the identified cell, it is possible that decreases in fluorescence intensity arise from reductions in activity within the surrounding neuropil. Thus, decreases in cellular activity should be interpreted with caution when attempting to assign a specific physiological function26. Nonetheless, the spatial and temporal characteristics of this IC strongly suggest that this signal is associated with dynamic Ca2+ activity of an individual cell. (b) Example of an IC with a lower signal-to-noise ratio than the example shown in a. The spatial filter shows morphological characteristics of a GCaMP-expressing cell; however, the activity trace shows a mixture of peaks that resemble those of temporal Ca2+ dynamics (red star), whereas others have nonrepresentative characteristics (purple star). The inclusion of ICs with low signal-to-noise ratio should be conducted with caution. Although this IC probably represents neural activity of a single cell, possibly with low activity levels, it may also be a partial component of a cellular signal or a noise artifact. (c) Example of an IC with morphological characteristics that weakly resemble a GCaMP-expressing cell (i.e., identified boundaries are not well defined). There is also a relatively high level of background fluorescence associated with this IC. Possible reasons for this weakly defined spatial filter may be that the IC represents an out-of-focus cell or possibly a cell whose shape has been optically distorted by the lens. Although the spatial filter of this IC is vaguely representative of a GCaMP-expressing cell, the activity trace shows temporal dynamics characteristic of Ca2+ transients. However, because of the weakly defined spatial filter, it would not be recommended that this signal be included for analysis. (d) It is possible that cellular components with highly correlated signals may be extracted as one independent signal. For example, the spatial filter shown here has morphological features resembling those of dendrites, possibly from more than one cell, and the corresponding activity trace has fluctuations in signal intensity vaguely resembling dynamic Ca2+ activity. Given that the spatial filter cannot be localized to one cellular component, it is not recommended to retain this IC for future analysis. Data shown here were collected over one imaging session from an adult male mouse expressing GCaMP6m (AAV-DJ-CamkIIa-GCaMP6m, titer: 7.0 × 1012, 1:1 dilution, Stanford) within the dentate gyrus. A 1-mm-diameter lens was used to visualize cellular activity. All procedures were approved by UNC IACUC. All scale bars, 50 μm.
Given that the intensity of Ca2+-encoded signals can change as a function of time, it is important to consider both the duration of an individual recording session42 and for how long a chronic imaging paradigm will be carried out29. More specifically, during an acute imaging session of particularly long duration (i.e., >40 min), it is possible for photobleaching of the signal to occur, resulting in a reduction in fluorescence intensity over time and/or a reduction in the total number of detected cells to decrease throughout the recording sessions. If signs of photobleaching are apparent, the experimental design should be modified to either reduce the total duration of an imaging session or modified so that imaging data are only collected at experimentally relevant time points. For example, for pharmacological manipulations, a 45-min imaging regimen can be used with 10-min intervals of the LED on, followed by a 5-min off period for a total of 30 min of imaging data42. The inclusion of LED-off periods into a relatively long imaging session will reduce the risk of photobleaching.
As described above, chronic imaging paradigms have an additional level of complexity, as steady increases in the expression of the Ca2+ indicator over time can result in both an increase in the fluorescence intensity of individual cells and the number of detected cells across sessions. Thus, during the experimental procedure, basal fluorescence intensity should be monitored to confirm that fluorescence emissions are stable throughout the duration of the experiment29. In addition to steady increases in fluorescence intensity, long-term expression of GCaMP may alter a cell's natural ability to buffer Ca2+ and therefore alter neural activity and/or behavior. To confirm that an observed change in neural activity is the result of a particular experimental manipulation and not merely due to the expression of GCaMP within a defined neural population, a control group that is imaged at similar time points as the test group, but does not undergo any other experimental manipulations, can be included (e.g., surgery positive/GCaMP positive/manipulation negative). Within the control group, neural activity should be stable throughout the duration of the test procedure. To confirm that the expression of GCaMP does not interfere with the performance of a behavioral task, the behavior of test subjects should be stable across time, or it should change in a manner that is predicted by the behavior of animals that have undergone similar experimental procedures but that do not express GCaMP (e.g., surgery positive/GCaMP negative/manipulation positive)42. Alternatively, this experimental group could express GFP within the target neuron population so that motion-induced changes in fluorescence intensity, as well as GCaMP-induced changes in behavior, can be controlled for in one group (e.g., surgery positive/GFP-positive/manipulation positive)35. Finally, experimental subjects can also be monitored for gross changes in behavior or general indicators of health42.
The experimental controls and precautions listed in this section are recommendations primarily based on the experience of our laboratory with this imaging methodology. Given the broad applicability of this technique to the field of neuroscience, these recommendations are unlikely to be a complete list. Thus, the appropriate experimental controls necessary for accurate data interpretation should be carefully considered before the start of an experiment. It is also important to consider whether a freely behaving design is the ideal method to answer the experimental question. For instance, although a freely behaving design may be appropriate for complex behavioral tasks that require an animal to be able to move naturally through its environment and appropriately adapt its behavior to a dynamically changing stimulus, such as during species-typical social behavior, it may not be ideal for experimental questions that require precise time-locked exposure to an environmental stimulus, such as studies related to sensory processing. In the latter case, if the goal is to conduct Ca2+ imaging within a deep brain neuronal structure, similar procedures for the introduction of GCaMP and the chronic implantation of a microendoscope may be used, as described in this protocol (Steps 1–47), and subsequently combined with head-fixed imaging procedures20,44.
Materials
REAGENTS
-
Mouse line of choice (wild type, Cre line for targeting specific cell types or transgenic mouse lines expressing GCaMP in a stable and uniform manner across cells for up to 6 months60 (available from the Jackson Laboratory, https://www.jax.org/))
Caution
The use of animal species should be conducted in accordance with institutional guidelines and regulations. The University of North Carolina Institutional Animal Care and Use Committee (UNC IACUC) approved the procedures used in this protocol.
-
Adeno-associated virus (AAV) constructs (see Table 1 for a list of available viral constructs). Our laboratory has successfully used GCaMP6 viruses available from the University of Pennsylvania, Stanford University and the UNC Vector Core. The data shown in the present paper are from viral constructs created at Stanford and UNC. Other groups have successfully used variants of GCaMP available from the University of Pennsylvania Vector Core with similar imaging procedures described in this protocol42,64
Table 1 Viral constructs for the introduction of a Ca2+ indicator into the brain. Caution
The use and disposal of AAV constructs should be conducted in accordance with institutional guidelines.
Critical
For long-term imaging studies, an alternative to viral delivery of GCaMP is to use transgenic mouse lines that stably express GCaMP.
-
Kwik-Sil adhesive (World Precision Instruments, cat. no. KWIK-SIL)
-
Kwik-Cast sealant (World Precision Instruments, cat. no. KWIK-CAST)
-
Mixer tips (World Precision Instruments, cat. no. 600009)
-
Super glue liquid professional (Loctite)
-
C&B-Metabond (Parkell, cat. no. S380)
-
Absolute ethanol, 200 proof (Fisher Bioreagents, cat. no. BP2818-4)
-
Isopropanol, 70% (vol/vol; Fisher Chemical, cat. no. A459-1)
-
Jet denture repair powder (Lang Dental, cat. no. 1250)
-
Ortho-jet crystal liquid fast curing acrylic resin (Lang Dental, cat. no. 0206; for pink dental cement)
-
Contemporary Ortho-jet liquid (Lang Dental, cat. no. 1506 Black; for black dental cement)
-
Acrylic primer (Lang Dental, cat. no. 1602)
-
Isoflurane
Caution
Gas anesthesia should be used in a well-ventilated room65
-
Medical-grade oxygen (Airgas, cat. no. OX USP125)
-
Topical anesthetic (Lidociane, USP (Akorn, cat. no. 17478-711-30)
-
Tissue adhesive (3 M; Vetbond, cat. no. 1469SB)
-
Eye ointment (Henry Schein, cat. no. 18582)
-
Sodium chloride, 0.9% (wt/vol) injection (Hospira, cat. no. 4888-20)
-
Betadine solution, poviderm medical scrub (Butler Schein, cat. no. 003659)
-
Ibuprofen, 160 mg/5 ml (Tylenol, Children's Tylenol)
EQUIPMENT
Surgery consumables
-
Ideal micro drill bur, 0.8 mm (Cell Point Scientific, cat. no. 60-1000)
-
Trephine for Microdrill (Fine Science Tools, cat. no. 18004-18)
-
Stainless steel disposable scalpels (Miltex, cat. no. 4-410)
-
5-0 Sofsilk sutures (Covidien, cat. no. D4G1516X)
-
Syringes, 1 ml (BD Biosciences, cat. no. 309659)
-
27G 0.5-inch blunt needle (SAI Infusion Technologies, B27-50)
-
27G 0.5-inch needle (BD Biosciences, cat. no. 305109)
-
25G 5/8-inch needle (BD Biosciences, cat. no. 305122)
-
18G 1-inch needle (BD Biosciences, cat. no. 305195)
-
18G 1.5-inch blunt needle (BD Biosciences, cat. no. 305180)
-
Dust-off compressed gas (Falcon Safety Products, DPSXL)
-
Gelfoam absorbable sponge (Pfizer Injectables, cat. no. 00009-0342-01)
-
12-well culture plates (Falcon, cat. no. 353043)
-
Skull screws (Glass and Watch Screws, 900 piece set, JT69900)
-
1/16-inch-diameter heat-shrink tubing (Qualtek, cat. no. Q2-F3X-1/16-01-MS100FT)
-
Carbon filter (ReFresh, cat. no. EZ-258)
-
Parafilm (Parafilm, PM-999)
Optical materials
-
Microendoscopic lens (see Table 2 for options available from Inscopix)
Table 2 Commercially available microendoscopes (from Inscopix). Critical
Methods in this protocol have been developed and tested using optical materials purchased from Inscopix. For other imaging system and microendocopic lens options, please see the following websites (http://doriclenses.com/, http://welcome.gofoton.com/).
-
Microscope baseplate (Inscopix, cat. no. BPL-2)
-
Microscope baseplate cover (i.e., dust cap; Inscopix, cat. no. BPC-2)
-
Microscope gripper tool (Inscopix, cat. no. GRP-1)
-
Hex key (Inscopix, cat. no. ATW-1)
-
Dummy microscope (Inscopix, cat. no. DMS-2)
-
Microscope objective lens cover (Inscopix, cat. no. MSC-2)
-
Optical cleaning kit (Fine Science Tools, cat. no. 29000-10)
-
Premium-grade lens cleaner (Thorlabs, cat. no. MC-50E)
Surgery tools and equipment
-
Vannas spring scissors (Fine Science Tools, cat. no. 15000-08)
-
Curved Graefe forceps (Fine Science Tools, cat. no. 11052-10)
-
Dumont 7 ceramic-coated forceps (Fine Science Tools, cat. no. 11272-50)
-
Dumont 5 forceps (Fine Science Tools, cat. no. 11251-10)
-
Dissecting chisel (Fine Science Tools, cat. no. 10095-12)
-
Curved 50-mm bulldog clamp (Fine Science Tools, cat. no. 18051-51)
-
Bonn micro-probe (Fine Science Tools, cat. no. 10033-13)
-
Micro-curette (Fine Science Tools, cat. no. 10082-15)
-
Bulldog serrefine (Fine Science Tools, cat. no. 18050-28)
-
Glass bead sterilizer (Simon Keller, cat. no. Steri 250)
-
Pump 11 Elite (Harvard Apparatus, cat. no. 70-4505)
-
7002KH 2.0-μl syringe (Hamilton, cat. no. 88400)
-
Modular routine stereomicroscope with 8:1 zoom (Leica, cat. no. M80)
-
Model 942 small animal stereotaxic instrument with digital display console (Kopf, cat. no. 942)
-
Non-rupture mouse earbar (Kopf, cat. ID Set 2, Model 922)
-
Electrode holder with removable open side clamp (Kopf, Model 1773)
-
Cannula holder (Kopf, Model 1776-P1)
-
Microdrill (Cell Point Scientific, cat. no. RS-67-1000)
-
Stereotaxic handpiece holder (Kopf, Model 1466-B)
-
Precision screwdriver (Westward, cat. no. 1UG41)
-
Homeothermic monitor (Harvard Apparatus, cat. no. 50-7220-F; for surgery)
-
Animal anesthesia kit (E-Z Anesthesia, cat. no. EZ-7000)
-
Oxygen regulator (E-Z Anesthesia, cat. no. EZ-320)
-
Heating pad (for recovery; Sunbeam, cat. no. 126982)
-
Heat gun (Weller, cat. no. 6966C)
-
Fixed sliding Probe Carriage (Scientifica, cat. no. PS-7750)
-
Patchstar micromanipulator (Scientifica, cat. no. PS07000C)
-
½-inch articulated ball and socket mount (Thor labs, TRB1)
In vivo imaging equipment and data analysis
-
SPRO ball bearing swivels with interlock snap (Cabelas, cat. no. IK-117351)
-
Model 923-B mouse gas anesthesia head holder (Kopf)
-
Arduino circuit board (Arduino, cat. no. A000066)
-
BNC (Bayonet Neill-Concelman Connector) (f) with American wire gauge (AWG) leads (Ponoma Electronics, cat. no. 4969)
-
Desktop with Intel core i7 processor, Windows 8.1, ideally with 32GB of memory (e.g., Lenovo, K450 Ideacenter; for data analysis)
Critical
In vivo Ca2+ imaging sessions create large data sets, and it is recommended to use a computer for the analysis of these data sets with specifications of at least an Intel core i7 processor, 8 GB of RAM, 2 TB free hard-disk space and USB 3.0 ports.
-
Desktop with Intel core i5 processor, Windows 8.1 and at least 8GB of memory (e.g., Dell, XPS8700; for data acquisition)
REAGENT SETUP
Virus preparation
-
Before stereotactic injection of the virus, if necessary, dilute the virus with sterile saline. The recommended virus dilutions are as follows: 1:2, 1:4, 1:8, 1:16 and 1:32. If the virus is being used that day, store it on ice until it is time to conduct the virus infusion procedure. Store all remaining aliquots at −80 °C. Under conditions of minimum freeze/thaw cycles, virus aliquots can typically be stored for ∼5 years.
Pink dental cement (for the foundation of the head cap)
-
Before the start of the surgery, fill each well of a 12-well plate with 2 g of acrylic powder. When you are ready to secure the microendoscope to the skull, mix 1 ml of clear Ortho-Jet liquid with the powder located in one well and load the mixture into a 1-ml syringe. Attach an 18G 1.5-inch blunt needle to the filled syringe. Wait at least 30 s before applying the dental acrylic solution to the skull.
Critical
Prepare well plates with dental cement before beginning surgical procedures for the implantation of a microendoscope.
Black dental cement (blocking background light and the attachment of the baseplate)
-
Prepare a well plate with acrylic powder, as described above. When you are ready to apply the black dental cement to the head cap or baseplate, add 1 ml of black Ortho-Jet liquid to the powder and load the mixture into a syringe, as described above. Wait ∼30 s before applying to the skull.
Critical
Prepare well plates with dental cement before beginning surgical procedures for the attachment of the baseplate.
Ibuprofen for postoperative care
-
Fill the water bottle that is to be placed in the test subject's home cage with 200 ml of water, and add 2.5 ml of Children's Tylenol (80 mg ibuprofen/200 ml water). Allow the water bottle to remain in the home cage for 2–3 d.
Postoperative recovery cage
-
Before the start of surgical procedures, it is recommended that a clean cage (fresh bedding, softened chow, nestlet material and a paper towel for the animal to be placed on immediately postoperatively) be prepared for the test subject to recover in. If a wire lid is used to hold chow, it is recommended that this be removed to reduce the number of physical objects in the cage that can potentially cause damage to the exposed surface of the lens. This analgesia and recovery procedure has been approved by UNC IACUC, and it should be conducted in accordance with the guidelines of your institution.
EQUIPMENT SETUP
Heat shrink–covered surgery tools
-
When you are handling delicate optical materials, such as the microendoscope, apply heat shrink tubing to hard metal tools. For general lens handling, apply heat shrink to the Dumont 5 forceps, with ∼10 mm of tubing added to each arm. Cut the heat shrink to an appropriate size, and fix the heat shrink to the forceps with the application of the heat gun (Supplementary Fig. 1a).
Lens holder
-
Cut two 5-mm-long heat shrink sleeves. Slide the heat shrink over each arm of the bulldog serrefine, and apply heat with a heat gun (Supplementary Fig. 1b). When it is time to implant the microendoscope, clamp the back end of the serrefine in the cannula holder (Supplementary Fig. 1c) and mount the entire apparatus on the stereotax (Supplementary Fig. 1d). Next, secure the microendoscope into the heat shrink–covered arms of the bulldog serrefine. When the lens holder has been secured to the stereotax, visually inspect the microendoscopic lens to ensure that it is level within the clamp. Once the microendoscope is level, it is ready to be lowered into the brain.
Critical
It is recommended that all lens handling procedures be conducted over the lens cloth to prevent damage to the lens in case it is accidently dropped during the preparation stage.
Bent needle for bone extraction
-
Use a curved graefe forceps to bend the tip of a 27G 0.5-inch needle ∼45 degrees (Supplementary Fig. 1e).
Sterile saline for irrigation during surgery
-
Attach an 18G 1-inch needle to a 1-ml syringe, and fill it with sterile saline. Use this syringe to apply sterile saline throughout the craniotomy and lens implantation procedure.
Procedure
Virus dilution study
Critical Step
If you are using transgenic mice expressing GCaMP in a stable and uniform manner across cells, this section is not necessary, and you can proceed directly to Step 5.
-
1
Before beginning in vivo imaging experiments, select the appropriate variant of GCaMP (fast (f), medium (m) or slow (s)) and optimize the concentration of the virus for the neuron population by first conducting a virus dilution study. Inject various dilutions of the desired viral construct into the brain region and mouse line of interest. Perform previously described stereotaxic procedures to introduce the viral construct into the brain (Fig. 4a).
Figure 4: Conduct virus dilution study. (a) Microinject GCaMP6 virus into the target neuron population. (b) Allow the virus to express for at least 3 weeks before examining the expression level of the indicator. (c–e) Confocal images showing GCaMP6f expression within the somatosensory (S1) cortex of a wild-type mouse (DIO-GCaMP6f; titer: 3.9 × 1012, UNC vector core). (c) 1:4 dilution shows overexpression of GCaMP6f in cortical neurons, as indicated by cells that lack a honeycomb appearance. (d) 1:8 dilution shows ideal expression of GCaMP6f, as there is a relatively large amount of healthy-looking cells expressing the GCaMP6f virus. (e) 1:16 dilution shows sparse expression of GCaMP6f. Ca2+ signals from such an expression are more difficult to detect compared with the 1:8 dilution, and they yield a low cell count.
Critical Step
Methods for virus introduction used in this step, such as microinjections with a Hamilton syringe or Nanoject, should be similar to those planned to be used in experimental subjects. Recommended virus dilution concentrations to test are 1:2, 1:4, 1:8, 1:16 and 1:32 of virus diluted from a high-titer viral stock.
Critical Step
Variants of the ultrasensitive GCaMP6 Ca2+ sensors outperform other sensors in vivo, but they still warrant consideration of temporal precision during live imaging and postprocessing. These indicators vary in kinetic properties and sensitivity, with sensors with slower kinetics having greater sensitivity. For example, GCaMP6f and GCaMP6m have faster kinetics (determined by the rise time + half the decay time) compared with GCaMP6s, and thus they have a greater potential to separate individual spikes within a burst or detect single action potentials in neuronal populations that exhibit fast firing rates. However, owing to the fast binding kinetics of these variants, GCaMP6f and GCaMP6m have lower signal intensities during excitatory events that are associated with a low number of action potentials14. Thus, selection of variant will depend on the firing rate and signal-to-noise ratio of the cell population being imaged.
-
2
Allow at least 3 weeks for sufficient viral expression to occur (Fig. 4b).
-
3
When sufficient time has elapsed for viral expression of the Ca2+ indicator, perfuse the subject as described in Wojtowicz and Kee66 and Li et al.67, and then examine the proportion of cells expressing GCaMP, as well as the health of GCaMP-expressing cells (Fig. 4c–e). Healthy cells should show nuclear exclusion of GCaMP6, and they should exhibit a honeycomb-shaped appearance (Fig. 5a,b and Supplementary Video 1). If GCaMP6 expression is observed throughout the entire cell body, resulting in a complete filling of the cell body with the fluorescence indicator, it is probable that overexpression of the Ca2+ indicator has occurred, which will lead to unhealthy cells and, eventually, to cell death (Fig. 5a).
Figure 5: Example of healthy and unhealthy GCaMP expression. (a) A neurotensin-Cre adult female mouse was injected with a 1:1 dilution of DIO-GCaMP6f (titer: 3.9 × 1012, UNC vector core) into the medial preoptic area (MPOA) and perfused 4 weeks post injection. 40× confocal image depicts the expression levels of GCaMP6f in MPOA neurotensin neurons. Arrowheads indicate cells showing nuclear exclusion of GCaMP6f (healthy expression), as well as nuclear filling (unhealthy expression). Owing to a relatively large number of cells showing nuclear filling of GCaMP at only 4 weeks, this dilution would not be recommended for this cell population. (b) Optical detection of healthy GCaMP-expressing neurons within the prefrontal cortex (PFC) of a freely behaving wild-type mouse. PFC neurons were visualized through a 1-mm-diameter GRIN lens with a head-mounted, miniature microscope secured to the skull of the mouse. The recording was conducted 8 weeks after virus injection (AAV-DJ-CamkIIa-GCaMP6s, 5.3 × 1012, 1:6 dilution, UNC vector core), and lens implantation was conducted over a 10-min session. A maximum intensity projection was made of the entire 10-min recording session to facilitate the visualization of optically detected neurons. In this image, neurons with dark centers can be visualized, which indicates a lack of nuclear filling and that optically detected cells are healthy. All procedures were approved by UNC IACUC. All scale bars 25 μm.
Critical Step
Critical Step
Optimal viral transduction is dependent on viral titers, as well as on the efficiency of a particular virus for transducing a given cell type. Caution should be taken when directly comparing viral titers between vector cores, as quality control assays and methods for determining physical and infectious titer can vary. Physical titer is expressed as the number of virus particles, viral genome or genome copies per unit of measurement. Functional or infectious titer is the measure of virus that infects a target cell. This is expressed as transduction units per ml or plaque-forming units per ml. These values are not absolute but rather a relative measure of infectivity, and direct quantitative measurements are therefore not comparable across vector cores.
-
4
If your experimental paradigm requires chronic imaging sessions (i.e., weeks to months), repeat Steps 1 and 2 with suitable viral dilutions, and examine viral expression at the longest time point planned for the experiment (e.g., 8–12 weeks after virus injection). It is particularly crucial that cell health be examined at these later time points to ensure that long-term expression of GCaMP does not result in cell death. Cell health can be examined after the completion of an experiment by examining mounted tissue sections under a microscope (i.e., post tissue histology)24,25 (Fig. 5a) or with one- or two-photon microscopy in living mice (awake or anesthetized) throughout the experiment29 (Fig. 5b). Post-tissue histology is recommended for the initial optimization of virus dilution, whereas one- or two-photon microscopy is best suited to confirm stable GCaMP expression and neural activity during the experiment.
Critical Step
It is normal for virus expression levels of GCaMP to increase stably and moderately over time, resulting in greater changes in fluorescence intensities compared with those measured at earlier time points, as well as an increase in the proportion of cells that express GCaMP17,27. Thus, it is crucial that stable virus expression throughout the duration of the study is optimized before the start of in vivo imaging experiments to ensure accurate data interpretation.
-
5
If you are targeting a genetically defined population of neurons, confirm cell type–specific expression using immunohistological methods (for example, Supplementary Fig. 2).
Selection of microendoscopic lens
-
6
Select the appropriate lens for your brain region of interest (Table 2 and Fig. 6a). The lens should be long enough to reach just above your neuronal population of interest, while still allowing for ∼2 mm of the lens to protrude above the skull (Fig. 6b). When selecting the appropriate microendoscope for your neuron population, it is also important to compare the optical properties of the microendoscope with the invasive nature of the lens implantation procedure. For example, both the transmission of light to and from the brain and the field-of-view size increase with lens diameter, which are optical properties that both affect signal detection capabilities (Supplementary Fig. 3). However, as lens diameter increases, so does the extent of tissue damage. Thus, the length and diameter of lens selected should be optimized to visualize the neuron population of interest, while minimizing damage to the underlying neural tissue.
Figure 6: Selection of microendoscopic lens and optimization of surgical placement. (a) Cartoon diagram of the microendoscopic lens (relay lens bound at either end by a GRIN lens) designed to transmit light below the surface of the brain. (b) Microendoscopic lenses of varying length and diameter are available for chronic brain implantation. Wide-diameter lenses (e.g., 1 mm) can transmit more light to and from the brain than lenses with narrow diameters (e.g., 0.5 and 0.6 mm), but the increased surface area of these lenses also results in more tissue damage. Thus, 1-mm-diameter lenses are best suited for more superficial regions of the brain, whereas 0.5-mm-diameter lenses that produce comparatively less pressure on the underlying tissue are better suited for deep brain regions. When selecting the appropriate microendoscopic lens, it is important to keep in mind that the total length of the lens must be sufficient to reach the target neuron population while still allowing for at least 2 mm of the lens to extend above the skull to accommodate dental cement application. (c) 10× confocal image showing an example of a correctly placed lens relative to the target neuron population. (d) 20× confocal image showing a lens that was placed medial to the target neuron population, resulting in a lack of GCaMP6-expressing neurons in the focal plane of the lens. For c and d, the tract for the microendoscopic lens is outlined in yellow, and the location of the focal plane is ∼290 μm below the bottom of the lens, which is outlined in orange. Tissue shown in both images was counterstained for DAPI (nuclear stain). All procedures were approved by UNC IACUC. Panel c is adapted with permission from Jennings et al.26.
Critical Step
Before beginning surgeries on experimental subjects, it is beneficial to optimize the placement of the microendoscope in relation to the target neuron population. For the lenses used in this protocol, the focal plane is ∼290 μm below the surface of the lower lens. Thus, to bring neuronal signals into focus, the bottom of the microendoscope should sit between 200 and 300 μm above GCaMP-expressing neurons (Fig. 6c). Given this relatively narrow range in the dorsal-ventral imaging plane, it is highly recommended that the viral indicator is also injected during the lens optimization procedure. During post-histology, the distance between the bottom of the lens and GCaMP-expressing neurons can be calculated to validate the appropriate placement of the lens (Fig. 6c). Optimizing these steps before beginning a large cohort of animals will decrease the number of subjects in which the focal plane of the lens is out of range of the target cell population (Fig. 6d).
Surgical preparation for craniotomy and lens implantation
Critical Step
Once the virus dilution has been optimized and the appropriate microendoscope has been selected, lens implantation surgery can begin on experimental subjects.
-
7
Before beginning surgical procedures, anesthetize the animal with isoflurane. Gas anesthesia is preferred over an injectable anesthesia, such as the combined use of ketamine and xylazine, because of the long duration of the surgery65.
-
8
Next, stabilize the mouse's head in the stereotaxic apparatus71 (Fig. 7a).
Figure 7: Surgery preparation for virus injection and microendoscopic lens implantation. (a) Stabilize the mouse in the stereotaxic apparatus. (b) Use forceps to pluck hair from the top of the skull. Digital image below shows example of how the skin should appear after a sufficient amount of fur has been plucked. (c) Perform alternating alcohol and Betadine scrubs (3×). Digital image below shows example of area of the skin where alcohol and Betadine should be applied. Betadine (brown liquid) is shown here. (d) Open the skin with a scalpel and bulldog clamps. Digital image below shows example of the size of the opening that should be made as well as appropriate placement of the bulldog serrefines. (e) Cartoon image showing the location of the bregma and lambda on the mouse skull. (f,g) Center the drill bit on bregma (f) and lambda (g) to level the skull. The distance in the z plane between the two locations should be less than 0.05 mm. (h) Level the skull in the medial-lateral direction by moving the drill bit to −2.0 mm posterior from bregma, and by measuring the difference in the z plane ±1.5 mm from the midline. The difference between the two points should be <0.05 mm. All procedures were approved by UNC IACUC.
-
9
Perform aseptic procedures in accordance with the guidelines of your institution. We perform surgical procedures in a designated, sanitized area (Fig. 7a); autoclave all instruments; pluck hair from the surgical site (Fig. 7b); and clean the surgical site with three alternating scrubs of isopropyl alcohol and Betadine (Fig. 7c).
-
10
To reduce the perception of pain at the surgical site and therefore to lower the level of general anesthetic needed to maintain anesthesia during surgery, we recommend application of topical lidocaine to the procedure site.
-
11
Make a small incision (∼12–15 mm) along the sagittal line of the skull by using graefe forceps to hold the skin taut while running a scalpel blade in a straight line from the anterior to posterior region of the exposed skin. Use four curved 50-mm bulldog clamps to further expose the skull (Fig. 7d). If necessary, use Vannas spring scissors to cut a larger opening in the skin, and remove any hair that may be too close to the area of the skull where the head cap will be made.
-
12
To increase the accuracy of stereotaxic procedures, ensure that the skull is level by comparing the height of bregma in relation to lambda (Fig. 7e). The difference in height between lambda and bregma should be less than 0.05 mm (Fig. 7f,g). If the difference in height between the two is greater than 0.05 mm, adjust the position of the skull in the dorsal-ventral direction until it is level. Next, ensure that the skull is level in the medial-lateral direction by measuring the height of the skull at a set distance posterior from bregma, such as −2.0 mm, and an equal distance lateral from the midline on either side of the skull, such as ± 1.5 mm (Fig. 7h). The difference in height at each location from the midline should also be less than 0.05 mm in the dorsal-ventral direction. If the difference is greater than 0.05 mm, rotate the mouse's skull using the bite bar, or reposition the mouse's head in the ear bar71.
-
13
To reduce the occurrence of movement noise during in vivo imaging sessions, remove connective tissue from the top of the skull and remove muscle near the top edge of the skull's surface. Use a cotton-tip applicator to remove periosteum and connective tissue from the skull's surface (Fig. 8a). Use the dissecting chisel to clear away all tendons and muscles located near the area of the skull where the head cap will be made72 (Fig. 8b). Apply sterile saline to the skull once all of the connective tissue has been removed.
Figure 8: Removal of connective tissue and muscle. (a) To reduce movement noise during imaging sessions, remove the connective tissue by rubbing a cotton-tip applicator on the top surface of the skull. Digital image below shows example of skull in which connective tissue has been removed has been removed by gently rubbing a cotton tip applicator on the top surface. (b) To further reduce the likelihood of movement artifacts, use a dissecting chisel to cut away muscle and tendons making contact with the top edge of the skull. Digital image below shows specific area of the skull where the dissecting chisel should be used to remove muscle. (c) When this step has been completed, the surface of the skull should be clear of connective tissue and the muscle should be retracted from the portion of the skull where dental cement will be applied during the head cap procedure (dotted outline on skull). All procedures were approved by UNC IACUC.
Critical Step
It is crucial that connective tissue, tendons, muscle, skin or fur are not allowed to come in contact with the dental cement that will be used to form the head cap (Fig. 8c). This can cause the head cap to become loose, thus increasing movement noise during recording sessions or possibly resulting in the loss of an experimental subject.
-
14
Use the 0.8-mm burr to drill holes for the placement of three skull screws (Fig. 9a). When drilling is complete, use an air can and a cotton-tip applicator to clear away any debris from the skull (Fig. 9a). To reduce the likelihood of excess bone fragments getting into the craniotomy site, we recommend that the drilling of skull screw holes and the removal of bone debris from this procedure occur before the craniotomy is performed.
Figure 9: Perform craniotomy. (a) Before performing craniotomy, drill three holes for the placement of skull screws in a triangle shape around the future craniotomy site. Clear away debris created by the drilling procedure (inset) before further opening up the skull to reduce the likelihood of bone debris entering the craniotomy. (b) Irrigate the skull with sterile saline to prevent tissue overheating, as shown in the cartoon in the digital image. (c) Center the trephine drill bit on bregma, and manipulate the drill using the stereotaxic arm so that the center of the trephine is located above the stereotaxic coordinates where the lens will be implanted. (d) Slowly lower the running drill to outline the craniotomy. Continue to drill until only a thin layer of skull remains. (e) Close-up of craniotomy outline. (f) Use a bent 27G needle to break away the remainder of the skull. Digital image below demonstrates where on the edge of the craniotomy the bent needle should be placed. (g) Once the center bone fragment is completely loose from the skull, carefully pull it away with a pair of fine-tipped forceps. Zoomed-in portion of g shows how the tissue under the fragment should appear. Notice that very little bleeding has occurred. (h) Irrigate the skull with sterile saline. Digital image below shows how much saline should typically be applied to the skull. All procedures were approved by UNC IACUC.
Critical Step
For maximum stability of the microendoscopic lens, each screw should be equally placed in a triangle shape about 1.5 cm from the craniotomy site (Fig. 9a).
-
15
Irrigate the skull with sterile saline to reduce tissue overheating during the craniotomy procedure (Fig. 9b).
Craniotomy, durotomy and tissue aspiration
-
16
Before performing the craniotomy, attach the trephine drill bit to the microdrill, and zero the center of the trephine drill bit above bregma (Fig. 9c). Move the trephine drill bit to the x,y coordinates where the microendoscope will be implanted (determined by mouse atlas)73.
-
17
To etch away the skull, slowly lower the trephine drill bit up and down on the exposed skull (Fig. 9d). Stop drilling when only a thin layer of skull remains (Fig. 9e). A craniotomy may also be performed using a 0.8-mm drill bit to etch a cranial window that is just large enough to perform the virus injections (if necessary), as well as lower the microendoscope. A cranial window that is just large enough to implant the lens will stabilize the lens and minimize movement noise.
Critical Step
To prevent overheating, do not allow the drill bit to remain in one location for too long. Overheating the skull may result in bleeding under the skull.
-
18
Use a bent 27G 1.5-inch needle (or Bonn microprobe) to break through the remaining skull, and loosen the large skull fragment in the center of the craniotomy (Fig. 9f).
-
19
With Dumont 7 ceramic-coated forceps, grab onto the large skull fragment and move it away from the brain (Fig. 9g).
-
20
Immediately irrigate the exposed tissue with sterile saline (Fig. 9h).
-
21
Use a bent 27G 1.5-inch needle (or Micro curette) to remove any remaining small bone fragments.
-
22
To facilitate virus injection and lens implantation, perform a durotomy (Fig. 10a). Use the bent 27G needle (or Bonn Micro Probe) to pierce through the dura, and slowly peel it away from the brain (Fig. 10b). Compared with brain tissue, dura has a yellow tint to it (Fig. 10b), and exposed neural tissue should appear white once the dura has been removed (Fig. 10c).
Figure 10: Durotomy, tissue aspiration and virus injection. (a) Perform durotomy to facilitate virus injection and lens implantation. Remove dura mater from the surface of the brain using a bent 27G needle. Hook dura on the tip of the needle and gently pull away from the brain. Continue to carefully pull away portions of the dura until a large enough opening is made for the injector needle and microendoscope to penetrate. (b) Example image showing the thin layer of dura pulled away from the brain tissue. (c) Image showing exposed brain tissue after completion of the durotomy. Brain tissue should appear white compared with dura mater. (d) Aspirate the tissue above the target structure with a blunt needle connected to a vacuum pump. (e) Aspirate in a circular motion while continually irrigating the tissue with sterile saline. Inset shows close-up of tissue before completion of the aspiration procedure. (f) Exposed brain tissue after aspiration of 1 mm of tissue. Inset shows close-up of tissue after aspiration. (g) If viral injection is required for the expression of the Ca2+ indicator, load an injector needle containing the virus into the stereotaxic electrode holder. (h,i) Center the injector needle on bregma (h), and, on the basis of stereotaxic coordinates for target neuron population, move the guide cannula to the region of interest and begin virus infusion (i). (j) When virus injection is complete, implant the skull screws. All procedures were approved by UNC IACUC.
Critical Step
When removing dura, be careful to avoid rupturing vasculature. If bleeding occurs during the procedure, pause the procedural step and apply a few drops of sterile saline to the exposed tissue. Use a pair of forceps to gently place a small piece of gel foam on top of the exposed tissue. Allow the saline-soaked gel foam to remain on the tissue until bleeding has subsided. Once bleeding has stopped, use a pair of forceps to remove the gel foam, and irrigate the area with sterile saline.
Critical Step
It is important that sterile saline is used in conjunction with the gel foam to prevent the gel foam from sticking to the tissue resulting in damage to the underlying tissue when the gel foam is removed.
-
23
Irrigate the exposed brain tissue with sterile saline immediately after completion of the durotomy.
-
24
Implantation of a microendoscopic lens deep within the brain will result in a large amount of pressure on the underlying neural tissue. To minimize tissue pressure, gently aspirate the tissue above your region of interest (at least 1–2 mm of superficial layers for deep brain regions; Fig. 10d). Before beginning the aspiration, bend a blunt 27G needle ∼45° and connect the bent needle to a vacuum pump. Test the vacuum pressure by slowly approaching the saline bubble on top of the skull with the needle, and adjust the pressure of the vacuum pump so that only a small volume of saline is aspirated at a time. To allow constant, fine control of suction pressure by hand, a 1-mm hole can be drilled into the plastic fitting of the needle providing an exhaust for re-direction of airflow20. Large changes in saline volume should not be immediately visible by eye. If large changes in volume are readily visible, the pressure of the pump should be lowered.
-
25
To begin tissue aspiration, slowly lower the needle toward the exposed neural tissue while continuously applying sterile saline, artificial cerebrospinal fluid (ACSF) or Ringer's solution for irrigation (Fig. 10e). Some bleeding is normal during aspiration, and adjustments in irrigation rates should be made to maintain tissue visibility, as well as to prevent the formation of large blood clots. This can be completed by stopping irrigation and aspiration for 5-s intervals, thus allowing only small clot formation. Gel foam can be used to control bleeding at the edges of the aspiration site. Aspiration should stop when at least 1 mm of tissue has been aspirated or at least a thin layer of tissue (500–800 μm) remains above your region of interest23. We have found more conservative estimates of tissue aspiration (i.e., aspiration of 1–2 mm of tissue when implanting a probe 4 mm into brain) to sufficiently reduce tissue pressure, while minimizing the risk of damage to the target structure (Fig. 10f). For some brain regions, structural landmarks can be used to determine when the aspiration procedure should stop. For example, when aspirating tissue for the implantation of a microendoscopic lens in the hippocampus, aspiration should stop when the white matter tracts of the corpus callosum become visible (indicated by a shiny appearance to the tissue).
Critical Step
We do not recommend that the tissue be aspirated for superficial structures, such as the cortex.
Critical Step
To further minimize pressure-induced tissue damage, a needle can be lowered about 2.0–2.5 mm above the region of interest and subsequently retracted. This needle serves as a way to further clear a path for lens implantation, and it does not function to suction tissue from the brain.
Introduction of Ca2+ indicator into neural population of interest
Critical Step
If the viral construct was injected at an earlier time point42,74, or if a transgenic mouse line that expresses a Ca2+ indicator is being used (http://www.jax.org), skip Step 26 and proceed straight to Step 27.
-
26
Use established stereotaxic procedures to introduce the selected GCaMP-encoding viral construct into the brain region of interest27,46 (Fig. 10g). To infuse virus in the mouse brain, our laboratory uses an injector needle connected to a 2-μl Hamilton syringe by PE20 tubing, but other methodologies for microinjections, such as with the use of a Nanoject, may also be used. Load the desired amount of viral construct (250–500 nl) into an injector needle, and secure the injector needle onto the stereotaxic apparatus (Fig. 10g). Center the injector needle above bregma, and move the needle to the appropriate stereotaxic coordinates (Fig. 10h,i). We recommend that the site of viral infusion be placed 250 μm medial-lateral and ventral to the site of lens implantation to minimize tissue damage under the lens (Supplementary Fig. 4). Infuse virus at a constant, slow rate of 100 nl/min. If you are imaging a vertical plane, such as with a prism lens that bends light at a 90° angle75,76,77, multiple injections along the imaging plane may be required. For example, if you are imaging within cortical regions of the brain, it may be necessary to administer at least three 250-nl injections spaced about 250–300 μm apart along the dorsal-ventral axis to successfully capture neural activity across multiple cortical layers. For 1.0-mm-diameter lenses, multiple injections along the medial-lateral axis (e.g., ±300–500 μm lateral from the center of the lens) may increase the amount of GCaMP-expressing cells in the focal plane, but it is not required for signal detection.
Lens implantation
-
27
Before beginning lens implantation, implant the skull screws in the holes drilled in Step 14 (Fig. 10j). These screws will act to anchor the dental cement to the skull. The final placement of the skull screws should not protrude out of the skull at a point higher than the final height of the implanted lens. This height will depend on the length of the lens that is being used, as well as the depth in which this lens is implanted in the brain.
-
28
Attach the prepared lens holder to the stereotaxic apparatus (Supplementary Fig. 1).
-
29
Prepare the microendoscope for placement into the lens holder by gently placing the microendoscope on the lens cloth from the optical cleaning kit. Use heat shrink–covered Dumont 5 forceps to pick up the microendoscope, and use an air can to gently remove any dust particles (Fig. 11a).
Figure 11: Implantation of microendoscopic lens for deep brain imaging. (a) Gently place the microendoscope onto the lens cloth and lightly grip it with heat shrink–covered forceps. (b) Image demonstrating stereotaxic setup for stabilizing the microendoscope during implantation. (c) Position the microendoscope above the stereotaxic coordinates for the target brain region and lower it into the brain at a constant, slow rate. (d) When you are done lowering the lens, place a small amount of Kwik-Sil around the lens to cover any exposed brain tissue. (e) Secure the microendoscope to the skull by carefully applying a thin layer of pink dental cement and letting it run to the lens and skull screws. Apply an additional layer of dental cement and let the cement dry for 15 min before removing the bulldog serrefine. (f) Dislodge the lens from the holder and cover head cap with black dental cement to block outside light. Build up the remainder of the head cap by forming a protective bowl around the lens with the black dental cement. (g) Apply Kwik-Cast to protect the top of the lens. All procedures were approved by UNC IACUC.
Critical Step
The microendoscope is composed of two GRIN lenses attached onto either side of a relay lens. To avoid damaging either imaging surface, the microendoscope must be handled with care.
-
30
Hold the microendoscope with heat shrink–covered forceps in one hand, and use the other hand to open the heat shrink–covered bulldog serrefine clamp. Place the microendoscope in the bulldog serrefine clamp and carefully adjust the placement of the microendoscope in the holder until it is level within the apparatus (Fig. 11b).
-
31
Without touching the microendoscope to the skull, zero the microendoscope just above bregma, and then move the microendoscope to the target location.
-
32
Before the lens is lowered, visually inspect the craniotomy site for bleeding.
Critical Step
It is crucial that all bleeding has stopped before the microendoscope is lowered, as excessive bleeding may occlude the field of view. If bleeding does occur at any stage of the procedure, apply gel foam to the craniotomy site until bleeding has subsided. If bleeding from the craniotomy site cannot be stopped, a lens should not be implanted as the bleeding will probably occlude the field of view under the lens.
-
33
Carefully lower the lens to ∼300 μm above the target neuron population. The microendoscope should be lowered at a constant, slow rate of ∼100–200 μm/min in steps of 10–20 μm (Fig. 11c). The slower rate of 100 μm/min is recommended to achieve optimal recording conditions. Bleeding should not occur during this process, and brain tissue, as well as small vasculature, should be visible under the lens throughout the duration of the lens implant procedure (Supplementary Fig. 5).
Critical Step
Lowering the microendoscope at a slow rate may provide the neurons, glia and vasculature time to separate, thus reducing the amount of tissue damage under the lens.
Critical Step
When you are lowering the microendoscope, retracting it up and down may also facilitate clearing a path for its implantation.
-
34
Once the microendoscope has been lowered to its final position, apply a small amount of Kwik-Sill around the lens within the craniotomy to cover any exposed tissue (Fig. 11d). It is not necessary that the amount of Kwik-Sil applied to the craniotomy exceeds the height of the skull. Application of too much Kwik-Sil may decrease the stability of the lens and therefore increase the likelihood of movement noise to occur during an in vivo imaging session.
-
35
To facilitate adherence of the dental cement to the skull, apply Metabond according to the application instructions provided in the kit suggested in the materials list. Alternatively, dental acrylic may be used, but Metabond is recommended.
-
36
Secure the microendoscope to the skull with dental cement. Use a 1-ml syringe connected to an 18G needle to apply a thin layer (∼1–2 mm thick) of the pink cement to the skull. Allow this layer of dental cement to run all the way to the microendoscope and skull screws (Fig. 11e).
Critical Step
This layer should be relatively viscous so that it can run to the lens on its own without any interference from the experimenter (i.e., manually pushing the cement toward the lens). This method will reduce the risk of bumping the lens during the dental application process.
-
37
Once the first layer of dental cement has dried (∼10 min), build up the head cap to cover the top of the skull screws (Fig. 11e). While the pink dental cement is still pliable, use a spatula to smooth the edges of the head cap, and move the dental cement away from any tissue. Allow this second application to dry for ∼15 min.
-
38
To further secure the microendoscopic lens to the dental cement, apply a small amount of super glue where the base of the dental cement meets the lens (Fig. 11e).
Critical Step
Avoid getting dental cement or super glue near the bulldog serrefine that is holding the microendoscope or on the exposed lens of the microendoscope.
-
39
Once the cement has completely dried, carefully dislodge the microendoscope from the bulldog serrefine. The microendoscope should not move when the bulldog serrefine is removed.
-
40
To prevent light other than that from the microscope-LED from entering the microendoscope, apply a layer of dental cement mixed with black acrylic around the head cap (Fig. 11f).
Critical Step
We do not recommend the use of black dental cement to adhere the microendoscope to the skull, as it does not adhere to the lens as efficiently as the pink cement used above.
-
41
To protect the lens from physical damage, use the black dental cement to form an additional ridge around the edge of the head cap that sits just below (∼1 mm) the top of the microendoscope (Fig. 11f). This ridge will also facilitate the application of a baseplate in Steps 65–73. The cement used in this step should be relatively thick so that the ridge is easily formed without the cement running to the microendoscope or onto the skin of the animal.
-
42
Smooth out the head cap with a spatula so that there are no sharp edges. Use scissors to clean up any excess dental cement. When the procedure is finished, the microendoscope should be secured to the skull without the head cap making contact with skin, connective tissue or muscle (Fig. 11f).
-
43
To prevent damage to the lens that could occur during daily activities in the home cage, place a layer of Parafilm over the surface of the lens and then generously apply a thick layer of Kwik-Cast to the top of the head cap, covering the entire surface of the microendoscope (Fig. 11g). The layer of Parafilm is recommended to prevent the Kwik-Cast from making direct contact with the lens. The layer of Kwik-Cast should be thick enough so that the mouse cannot easily scratch through it.
-
44
Disengage the mouse from the stereotax and discontinue anesthesia.
-
45
Allow the mouse to recover in a clean cage placed on top of a heating pad until it is ambulatory.
-
46
Administer postoperative analgesia according to the guidelines of your institution. We use Children's Tylenol suspended in the home cage water bottle for 2–3 d after surgery. Allow 3–6 weeks for sufficient viral expression to occur and for the tissue to heal before proceeding with the next step.
Checking Ca2+ indicator expression and attachment of baseplate
Critical Step
Imaging experiments can begin when GCaMP is sufficiently expressed, in order to allow for the detection of neural activity under the lens with a miniaturized epifluorescence microscope. The specificity of this time point will depend on the brain region and cell type. In general, we have found that distinct Ca2+ transients can be detected from individual cells within cortical regions at an earlier time point than deep brain target structures. On average, after virus injection and lens implantation procedures, Ca2+ transients can be distinctly visualized as soon as 2 weeks for cortical regions and 3–6 weeks for deep brain regions. Procedures for checking the expression levels of GCaMP are described in Steps 47–64.
-
47
When enough time has elapsed for sufficient expression of GCaMP (∼2–6 weeks), use the miniature epifluorescence microscope to check for Ca2+ signals, as well as to optimize the placement of a baseplate that will act as a docking station for the microscope.
-
48
While holding the microscope, adjust the focusing mechanism on the microscope so that it is in the mid-range (about 1 and a half turns up from the lowest starting point) (Fig. 12a).
Figure 12: Checking the expression of the Ca2+ indicator. (a) Interface the miniature, integrated microscope with the acquisition computer by connecting the microscope with the baseplate attached to the DAQ box and the DAQ box to the acquisition computer. The blue arrowhead in the inset points to the set screw on the microscope focusing mechanism, which locks the focus in place. (b) Secure the mouse in the stereotaxic apparatus. (c) Remove the Kwik-Cast from the top of the head cap and clean the lens with 70% (vol/vol) ethanol and lens paper. (d) Align the objective of the microscope over the top lens of the microendoscope. The two lenses should be ∼2 mm apart and parallel to each other. (e) Turn on the LED to ensure that the center of the LED is passing through the center of the microendoscope. (f) Visualize the objects under the lens using the acquisition software. The first object to come into focus is the top of the lens. All the edges of the top lens should be clear and crisp if the microendoscope is level with the objective of the microscope. (g) As you move closer to the top of the lens, the vasculature and fluorescence signals will begin to come into focus. (h) The vasculature and outlines of cell bodies should be clear when in focus. (i) Example images of biological structures under the lens, 3 weeks (top) and 5 weeks (bottom) after surgery. If cell bodies and vasculature outlines are not yet clearly defined, it is ideal to wait to attach the baseplate. If the outlines of cell bodies and vasculature are clear and crisp, attach a baseplate to the head cap of the animal. Scale bars, 250 μm. All procedures were approved by UNC IACUC.
Critical Step
Attaching the baseplate while the microscope is in the mid-range of its focusing potential will maximize the range that the microscope can be focused in either direction.
-
49
Secure the focusing mechanism in the mid-range position by tightening the set screw on the focusing mechanism (Fig. 12a).
-
50
Next, turn the set screw on the baseplate until it is flush with the edge of the baseplate, and then attach the baseplate to the microscope by turning the set crew on the baseplate from 0.25–0.5 turns (Fig. 12a).
-
51
Attach the microscope gripper tool to a micromanipulator, and then secure the microscope in the gripper tool to allow the microscope to be accurately maneuvered above the implanted microendoscope. We recommend the Patchstar micromanipulator, but an articulated ball and socket mount from Thorlabs may also be used, as well as any other brand of quality micromanipulators.
-
52
Connect the data acquisition (DAQ) box to the imaging computer with a USB 3.0 cord (Fig. 12a).
-
53
Connect the microscope to the DAQ box (Fig. 12a).
-
54
Position the stereotax under the microscope that has been secured to the microscope gripper tool. The microscope should be centered above the earbars so that the microscope is above the head of the mouse when it is secured in the stereotax.
-
55
Anesthetize the mouse (we use isoflurane anesthesia)65 and secure it in the stereotaxic apparatus (Fig. 12b).
-
56
Use fine forceps to remove the Kwik-Cast from the top of the microendoscope (Fig. 12c).
Critical Step
Be careful to avoid the area underneath the Kwik-Cast where the microendoscope has been implanted.
-
57
Use compressed air to clear any debris from the top of the microendoscope.
-
58
Gently clean the exposed lens of the microendoscope with lens paper and 70% (vol/vol) ethanol.
-
59
Use the micromanipulator to center the objective over the microendoscope so that it is within ∼2 mm of the top of the lens (Fig. 12d).
-
60
Level the objective of the microscope relative to the microendoscope by adjusting the microscope position in the gripper tools in the medial-lateral direction and the head of the mouse in the anterior-posterior direction.
-
61
Once the surface of the microscope lens objective is level with the implanted microendoscopic lens, start the imaging acquisition software and use it to adjust the settings on the microscope and to visualize cellular activity (Fig. 12d).
-
62
Turn on the LED of the microscope and make sure that the light path of the LED is going through the center of the lens (Fig. 12e).
-
63
Adjust the intensity of the LED, gain and frame rate to visualize Ca2+ activity. Optimal LED intensity and gain will vary by lens properties, brain region, cell type, as well as by the properties of the Ca2+ indicator, such as sensitivity, brightness, Ca2+ binding kinetics and labeling efficiency13,19.
Critical Step
To avoid photobleaching of fluorophores or phototoxicity, it is recommended that the intensity of the LED be set below 20% of its maximum power. However, it is important to keep in mind that light output can vary across microscopes and that the amount of light that penetrates the brain will also depend on the diameter of the lens, as larger-diameter lenses will allow more light to be transmitted through the lens. Thus, to determine the output of light that reaches the focal plane below the lens (∼290 μm below the surface of the lens), the output of the microscope LED through the GRIN lens would need to be measured for each system. It has previously been reported that the transmission of 0.05–0.20 mW of light through a GRIN lens results in virtually no photobleaching over a 20-min imaging session29, but this may vary on the basis of the exact preparation. The effect of LED outputs above this range or recording duration on fluorescent signals would need to be tested empirically for each experimental setup.
-
64
While the LED is on, use the micromanipulator to approach the surface of the lens. As you approach the lens with the micromanipulator, the top of the lens should come into focus. If the objective is level over the lens, all edges of the lens should be clear and crisp (Fig. 12f). As you continue to lower the microscope close to the top of the lens, the tissue should begin to come into focus, as indicated by diffuse fluorescence signals resulting from out-of-focus light, as well as the faint outlines of vasculature (Fig. 12g). When the tissue has fully come into focus, examine the quality of tissue in the field of view. The vasculature should be clearly visualized, and the outlines of GCaMP-expressing cells should be distinct (Fig. 12h). If you are checking cells at an early time point (e.g., 2–3 weeks after surgery) and fluorescence signals are low, follow option A. If distinct GCaMP-expressing cells can be visualized, follow option B. If fluorescence signals have the appearance of autofluorescence, follow option C. If the visual field is black, follow option D. If cells can be visualized but little neural activity is seen, follow option E. If detectable Ca2+ signals do not appear 8–12 weeks after surgery, follow option F.
-
A
If low fluorescence levels are visualized at an early time point
-
i
If there are dynamic fluctuations in Ca2+ activity, but the fluorescence levels are low or only bulk changes in fluorescence can be visualized, it may be beneficial to wait 2–3 more weeks before attaching the baseplate. Allowing more time for the expression of GCaMP may facilitate the visualization of distinct cellular signals and therefore optimize the placement of the microscope objective relative to the implanted microendoscope.
-
ii
In this scenario, re-apply Kwik-Cast to the lens, and then return the animal to its home cage.
-
iii
Repeat Steps 48–65 at a later time point. We have found 5–6 weeks after virus injection and lens implantation to be an optimal time point for the attachment of the baseplate in most of our preparations (Fig. 12i).
-
i
-
B
If distinct GCaMP-expressing cells can be visualized
-
i
Confirm that there are dynamic changes in fluorescence activity indicative of Ca2+ transients by switching to the ΔF/F function on the software. If fluorescence signals are representative of Ca2+ transients, you should see dynamic changes in fluorescence activity. The dynamic changes in signal intensity should resemble the morphology of cells (i.e., spherical, see Fig. 3) and should not correlate with tissue movement.
-
ii
Proceed to Step 65.
-
i
-
C
If autofluorescence or cell death is detected
-
i
If cells have died because of overexpression of the Ca2+ indicator (Supplementary Fig. 6), or if fluorescence signals have the appearance of cells but are actually autofluorescence from tissue damage, dynamic changes in fluorescence activity will not be visualized and a baseplate should not be attached (Fig. 13).
Figure 13: Optically detected signals that are not indicative of fluorescently encoded Ca2+ transients. (a) Tissue damage or cell death can result in the production of fluorescence signals, referred to as autofluoresence, at the site of injury. (b) These autofluorescence signals can be detected with a single-photon fluorescence microscope and imaged through a microendoscopic lens. Importantly, aggregates of autofluoresence signals may display similar morphological characteristics of cells (i.e., spherical spheres of fluorescence). However, unlike GCaMP6-expressing cells, signals encoding autofluorescence lack dynamic changes in signal intensity, allowing these signals to be easily distinguished from those representative of neural activity. (c–d) Application of an automated cell sorting algorithm (PCA/ICA) to the data shown in b failed to extract signals that were representative of dynamic cellular activity. Instead, only background fluorescence (c) or biological structures (d), such as blood vessels, could be extracted from the data. (e) Activity traces extracted from background fluorescence signals shown in c lack characteristic temporal dynamics of fluorescently encoded Ca2+ signals, further indicating that it is unlikely that the fluorescence signals detected are representative of dynamic Ca2+ activity. It is not recommended to attach a baseplate if fluorescent signals have the appearance of autofluorescence. All procedures were approved by UNC IACUC.
-
ii
Perfuse the subject by intravascular perfusion with PBS followed by paraformaldehyde66,67. Remove the brain and postfix it in PFA for 24 h. After postfixation, transfer the brain to 30% (wt/vol) sucrose in PBS for cryoprotection, and keep the brain in sucrose solution until it sinks to the bottom (∼24–48 h)78. In order to preserve the lens track in tissue, the lens can be left in the brain for 3–4 additional days while in PFA.
-
iii
Section perfusion-fixed tissue with a cryostat78 and examine the quality of tissue near the lens, as well as viral expression of GCaMP, with an epifluorescence microscope79. A large amount of tissue damage near the lens track may indicate that the lens was lowered into the tissue too quickly (Fig. 13a).
-
i
-
D
If the visual field is black
-
i
If the visual field under the lens is completely black, it is possible that the transmission of light is being occluded by physiological properties within the brain, such as a blood clot.
-
ii
Perfuse the mouse (as described in Step 64C(ii)) and visually examine the tissue below the lens.
-
i
-
E
If anesthesia suppresses neural activity
-
i
Neural activity is often dependent on the conscious state of the animal, and it may be suppressed if the animal is deeply anesthetized. Gently pinch the tail of the animal or make a loud noise, such as a handclap, to try and stimulate neural activity and to facilitate the visualization of Ca2+ dynamics.
Critical Step
It is possible that exposure to certain types of anesthesia may suppress the activity of specific neuronal populations, resulting in an inability to visualize dynamic changes in fluorescent activity. Although we have yet to identify a population of neurons that could not be visualized within an anesthetized subject, if a neuron population is sufficiently suppressed by isoflurane to prevent its visualization, the level of anesthesia could be temporarily lowered or another type of anesthesia, such as ketamine and xylazine, could be used65.
-
i
-
F
If there are no detectable Ca 2+ signals after 8–12 weeks
-
i
Perfuse the mouse as described in Step 64C(ii).
-
ii
Check the lens placement position relative to viral expression, as described in Step 64C(iii).
-
i
-
A
-
65
Move the position of the microscope relative to the microendoscope to select the focal plane that contains the greatest amount of healthy cells, and turn off the LED.
Critical Step
Only proceed with this step if cells appear healthy and a sufficient level of GCaMP expression was identified in Step 64.
Critical Step
Some adhesives may exhibit shrinkage during the drying process, resulting in a slightly lower baseplate position than intended. To minimize improper positioning of the baseplate owing to shrinkage, test your adhesive before the baseplate attachment procedure to ensure that a substantial amount of shrinkage does not occur. Alternatively, the baseplate may be secured slightly above the desired focal plane to compensate for movement that may occur during the drying process.
-
66
Use black dental cement to permanently secure the baseplate to the head cap. Fill a 1-ml syringe with dental cement and attach a blunt 18G needle to the end. The dental cement should be thick and easily malleable.
-
67
Begin attaching the baseplate to the head cap by building four pillars, one at a time, running from each corner of the baseplate to the head cap (Fig. 14a,b).
Figure 14: Attachment of the baseplate. (a) Begin attaching the baseplate (outlined with white dotted line) to the head cap by forming pillars of black dental cement from the baseplate to the head cap. (b) Form four pillars in total, one at each corner of the baseplate. (c) Build up all remaining walls of the head cap (edge of dental cement outlined in green). (d) Before building up the final posterior wall, turn on the LED (i) and make sure that no light escapes from the completed walls (ii). Fill in any cracks from which light escapes, and complete the baseplate attachment procedure by filling in the posterior wall. (iii) Once all walls have been filled, cells and vasculature that were visible when the focal plane was selected should be visible when the LED is turned on. Scale bar, 250 μm. (e) Example image of anterior and lateral wall of the head cap with the dental cement going about half way up the baseplate. (f) Example image of the posterior wall and lateral wall of the baseplate where the set screw is located. The posterior wall of the baseplate is shorter than the other three walls, so caution should be taken when applying dental cement to this region. Caution should also be taken when applying dental cement near the set screw of the baseplate or when in close proximity to the microscope circuit board. (g) Before removing the microscope, test to make sure that the dental cement has completely dried (∼15 min) by scratching the exterior with a spatula. This should make a scratching sound once the cement has hardened. (h) Once the dental cement has dried, dislodge the microscope from the microscope gripper. (i) Use the hex key to loosen the baseplate set screw. (j) Remove the microscope by gently pulling up on the microscope while applying slight downward pressure to the mouse's head. (k) Secure the dust cap on the baseplate to protect the top of the lens by locking the dust cap in place with the set screw. All procedures were approved by UNC IACUC.
-
68
Begin slowly building up dental cement from the head cap to the baseplate by filling in the four walls surrounding the baseplate (Fig. 14c). Build up the anterior and two lateral walls first.
-
69
When the anterior and lateral walls are complete, turn on the LED and make sure that no light escapes from these three walls (Fig. 14d). If any light escapes, fill in cracks with dental cement.
-
70
Turn off the LED and build up the posterior wall.
Critical Step
The posterior wall of the baseplate is shorter than the other walls, so extra caution should be used when applying dental cement to avoid getting dental cement on the microscope.
-
71
Before completing the dental cement application, inspect the baseplate to make sure that there is a sufficient amount of dental cement on all four walls of the baseplate so that it is securely attached to the head cap (about half way up the baseplate) (Fig. 14e,f).
Critical Step
Be careful to avoid getting dental cement on the microscope or near the set screw located on the baseplate (Fig. 14f). Because the space between the baseplate and head cap is extremely narrow, it may be difficult to apply the dental cement without getting it on the microscope or near the lens. The dummy microscope can be used to practice the baseplate surgery until one feels comfortable with the procedure. To do this, attach the baseplate to the dummy microscope and the dummy microscope to the holder, as described in Steps 50 and 51. Next, place a glass slide under the microscope and practice attaching the baseplate to the slide (Supplementary Fig. 7).
Critical Step
Improper baseplate attachment is the leading cause of motion artifacts, and achieving optimal viscosity of the dental cement is crucial to proper baseplate attachment. If the dental cement is too runny, it is possible that it will flow to the lens and cover up the imaging surface. If the cement is too solid, it will not properly adhere to the baseplate, thus increasing the probability of movement noise.
-
72
Once the dental cement has completely dried (Fig. 14g), dislodge the microscope from the gripper tool and move the micromanipulator arm away from the microscope (Fig. 14h). The microscope should remain stable on the mouse's head during this process.
-
73
Use the hex key to loosen the set screw on the baseplate (Fig. 14i). Gently pull the microscope away from the baseplate while holding the mouse's head down with the other hand (Fig. 14j).
-
74
Carefully place the microscope back in its case.
-
75
Attach the dust cap to the baseplate. As above, turn the set screw on the baseplate so that it is flush with the baseplate. Attach the dust cap to the baseplate and tighten the set screw 0.25–0.5 turns until the dust cap is securely attached to the baseplate (Fig. 14k). Allow the mouse to recover for 2–3 d before beginning habituation procedures or behavioral testing.
Preparation for in vivo monitoring of neural activity in freely behaving mice
-
76
Although the microscope is extremely lightweight (∼2 g), some mice take time to adjust to the attachment process and initially have difficulty maneuvering through their environment with the microscope attached. If it seems necessary, habituate the mouse to these procedures, by using a dummy scope secured to a tether with similar physical and mechanical properties as the microscope cable (Supplementary Fig. 8a), and place the mouse in a behavioral testing setup that will be used during the experimental procedure42,74 (Supplementary Fig. 8b). When the mouse appears to be moving normally through its environment (i.e., unrestricted active forward locomotion and turning behavior), proceed to the next steps to begin in vivo imaging experiments.
-
77
Make sure that the computer used for imaging has enough space to store the data. For example, 30 min of data acquired at 20 Hz require ∼110 GB of space.
Critical Step
Check this before beginning a behavioral recording session to avoid data loss.
-
78
Connect the microscope imaging system to the acquisition computer, as described in Steps 52 and 53.
-
79
To attach the microscope, anesthetize the mouse with isofluorane.
-
80
Place the mouse on a heating pad with its nose positioned in front a rebreathable nose cone apparatus to maintain gas anesthesia (Fig. 15a).
Figure 15: Visualization of Ca2+ transients in freely behaving mice. (a) Anesthetize the mouse and remove the dust cap. (b) Mount the microscope to the baseplate. (c) Adjust the focusing mechanism to bring cell bodies and vasculature into focus. (d) Tighten the set screw on the focusing mechanism once the focal plane has been selected. (e) After anesthesia recovery period, conduct Ca2+ imaging during naturalistic, unrestrained behavior. (f) 2D xy image stacks of raw Ca2+ imaging data acquired during an in vivo imaging session. Scale bar, 250 μm.
-
81
Remove the lens cover from the microscope, and use the hex key to remove the dust cap from the baseplate (Fig. 15a).
-
82
Turn the set screw on the baseplate so that it is flush with the inner wall of the baseplate, and attach the microscope to the baseplate (Fig. 15b) by turning the set screw on the baseplate 0.25 to 0.50 turns.
Critical Step
It is essential that the set screw for the focusing mechanism or baseplate are never overtightened. When securing the microscope or the focusing mechanism in place, both screws should never be turned more than 0.25–0.5 turns. Turning of the screw should be stopped as soon as any resistance is felt. Overtightening the set screw may result in the microscope becoming dislodged during the recording or possibly damaged.
-
83
Start the data acquisition software. Adjust the frame rate, LED intensity and gain to visualize cellular activity. As described in Step 63, it is best to keep the intensity of the LED between 10 and 20% of its maximum power to avoid photobleaching or cell death as a result of phototoxicity. If the signal still needs to be amplified after this point, compensate for a low signal by either increasing the exposure time (i.e., the length of time in which light is collected for a given frame), or by adjusting the gain.
-
84
Adjust the focusing mechanism on the microscope to increase or reduce the distance of the sensor from the focal plane until cells and vasculature are clearly in focus (Fig. 15c).
-
85
Once the focal plane has been selected, lock the focusing mechanism in place with the set screw (Fig. 15d). Allow the mouse to recover with the microscope attached to its head in a secure cage for at least 20–45 min before beginning behavioral recording session (Fig. 15e)29,42.
Critical Step
The use of anesthesia, even over brief periods, may result in long-term suppression of neural activity. Confirm recovery from anesthesia by observing normal animal movement and a return to baseline levels of neural activity. It may also be beneficial to conduct recordings of the target cell population as the mouse recovers from anesthesia to determine the amount of time it takes for anesthesia-induced changes in neural activity to wear off. Finally, to control for any remaining anesthesia-induced changes in neural activity, it is important that all mice are given the same amount of time to recover from anesthesia before beginning a recording session. If the use of anesthesia substantially disrupts your experimental design, it is possible to tether the mouse by having one person manually restrain the mouse while another person removes the dust cap and secures the microscope. However, the use of manual restraint to attach the microscope also has its limitations, as it is likely to induce stress in the test subject. For this reason, it is important to habituate the test subject to the attachment procedure and to allow the mouse sufficient time to recover. It is also important that all test subjects undergo similar microscope attachment procedures.
-
86
When sufficient time has elapsed to allow the mouse to fully recover from anesthesia, place the mouse in the behavioral recording chamber.
-
87
If the microscope that you are using does not allow for the use of a commutator, secure the microscope cable at a point high above the floor of the chamber where the recording session is to be conducted (∼120 cm). Securing the microscope cable at a distance high above the base of the recoding chamber will reduce the amount of twisting in the cable that often occurs near the top of the microscope. In addition, attaching the cable to an object with the ability to stretch, such as a rubber coil, or to a small object with a turning mechanism, such as a ball-bearing swivel, will further reduce strain on the cable and facilitate animal movement.
-
88
Once the cable is secure, ensure that there is sufficient slack in the cable to allow the animal to freely roam about the testing chamber. Given that both the microscope and attached cable are light enough to be carried on the head of the mouse without the necessity of external support, an optimal configuration of the tether provides enough slack so that the mouse can freely move about its environment without the tether hanging below the head of the mouse. If the cable is too tight, it will result in excessive strain on the cable, possibly resulting in damage to the thin copper wires located within the cable and a subsequent loss of data. Alternatively, if the cable is too loose, the animal may be able to access it and bite through the thin wires, Figure 15e.
-
89
Open the acquisition software and select the frame rate that is optimal for your neuron population and indicator (Fig. 15f). Typically, it is ideal to select the variant of GCaMP (i.e., fast, medium or slow) on the basis of the physiological properties of the specific neuronal population, and to select the frame rate on the basis of the kinetic properties of the indicator.
Critical Step
It is important to keep in mind that a higher frame rate will reduce the intensity of the detected signal (indicated by a lower mean pixel intensity) as a result of a reduction in exposure time. Thus, to reduce the possibility of photobleaching, it is best to not record at a frame rate that is higher than necessary to accurately detect the neuronal signal. In addition, higher frame rates increase the amount of acquired data, potentially leading to unnecessarily high data storage cost, as well as an increase in data analysis time.
-
90
Next, turn on the LED and adjust the LED intensity and gain to maximize the fluorescence signal. If provided by your acquisition software, use the mean intensity pixel value feature to facilitate the selection of LED intensity and gain. To achieve pixel values within the recommended intensity range of 1,500–3,000, first adjust the LED intensity to up to 20% of its maximum intensity. Adjusting the LED intensity produces the least amount of noise when enhancing signal detection.
-
91
If signal intensity is not sufficiently high, adjust the gain to digitally enhance signal brightness. Keep in mind that using the gain to adjust signal intensity will also increase noise, so it is recommended that gain be kept at 1 when possible. A threshold indicator feature may be used to ensure that the LED and gain are not so high that they can result in signal clipping or photobleaching/photoxicity.
Critical Step
We have found that a mean intensity pixel value ∼3,000 works well for most recording regions and neuron populations. It is recommended that the mean pixel intensity does not fall below 1,500. A pixel value below this range may make it difficult to extract significant cellular signals from the acquired images. Conversely, a mean pixel value above 4,000 may overexpose the tissue, resulting in photobleaching or phototoxicity.
Critical Step
Long-term recording sessions (i.e., greater than 20 min) have an increased risk for photobleaching, photoxicity and potential tissue damage because of heating of the tissue by the LED. It is recommended that frame rate, LED and gain settings be adjusted as described in this protocol to minimize photobleaching. Although tissue heating has not been directly tested (or reported) in response to exposure to light produced by LEDs located within miniaturized microscope systems, prior studies using microinorganic LEDs directly implanted in brain tissue have reported LED-induced increases in tissue temperature as a function of stimulation frequency over time80. However, we have measured the output of light directly from the objective lens of the microscope and found that light output produced by the LEDs located in miniature microscopic systems falls well below the range of light intensities associated with heating of neural tissue80,81. Thus, we predict that tissue heating due to the LED should be negligible.
-
92
For experiments that require multiple recording sessions, a snapshot of the imaging site can be taken on the first imaging session to facilitate repeated imaging of the same field of view. If appropriate, take such an image. On future imaging sessions, the snapshot can then be uploaded into the imaging software and overlaid on top of the live image to ensure that the biological structures underneath the lens appropriately align. For example, if using nVista recording software, this can be accomplished by selecting overlay options under visualization tools and uploading a TIFF file that is representative of the focal plane from the previous recording session. Next, adjust the opacity of the TIFF image so that the live image can be visualized underneath the TIFF image. The focal plane can then be adjusted with the microscope focusing mechanism until the location and size of anatomical structures is consistent between recording sessions. In addition, the number of turns that the focusing mechanism was adjusted from its lowest point can be counted by placing evenly spaced tick marks around the top of the focusing mechanism. An example of such tick marks can be seen in Figure 15c,d. By combining both suggested methods, we have successfully imaged the same neuronal population across multiple imaging sessions in one mouse (Supplementary Fig. 9a,b). We further maximized repeated cell identification by applying an image-registration algorithm to the extracted spatial maps (Supplementary Fig. 9c–e).
-
93
Allow the mouse sufficient time to recover from anesthesia and to habituate to the recording session.
-
94
Immediately before beginning an imaging session, check the settings of the gain and the LED to ensure that the settings are optimal. Although the intensity of the LED and gain may have been adjusted to visualize cells while adjusting the focus of the microscope, it is common for anesthesia to suppress neural activity, and thus the settings used to initially visualize the cells may be higher than those needed for signal detection in an awake freely behaving animal. If needed, adjust the settings.
-
95
For experimental procedures that do not require the acquisition software to be triggered by an external program, follow option A. For experiments that require the acquisition of imaging data to be synchronized with a behavioral acquisition system, follow option B.
-
A
For non-synchronized recordings
-
i
Adjust the recording schedule in the acquisition software to automatically set the duration of the recording session.
-
ii
Select 'Record' in the imaging acquisition software to begin an imaging session.
-
i
-
B
To synchronize imaging data with behavioral monitoring software
-
i
To trigger an imaging session via an external program, connect the BNC output cable from the behavioral recording system to the trigger input port on the DAQ box. If you are using the nVista imaging system, the DAQ box contains a BNC port labeled 'TRIG' that allows it to receive signals to trigger the start and stop of an imaging session from another software.
-
ii
Press 'start' on your designated behavioral recording software to trigger the imaging acquisition software.
Critical Step
In our experience, behavioral recording systems that send low-output voltage signals work best when integrating the DAQ box with behavioral acquisition software. If your system sends a high-output signal, an Arduino microcontroller or a simple circuit using a transistor can convert this signal to a low-output signal (and maintain the base state at high).
Critical Step
We have also found that it is best to run the imaging acquisition software and the behavioral acquisition software on separate computer systems.
Critical Step
A recording session can also be triggered by coupling the acquisition software with commonly used behavioral recording software, such as VideoTrack (ViewPoint Behavior Technology)42, Ethovision (Noldus)26 or Med Associates behavioral equipment26. Triggering the microscope in this manner will allow the imaging data to be synchronized with behavioral data.
-
i
Critical Step
For both options A and B, it is important to consistently monitor the animal and cable throughout the duration of the imaging session. In addition, while imaging, nonessential applications on the computer should be closed to reduce the risk of the data acquisition crashing during a recording session.
-
A
-
96
Once the imaging session is complete, anesthetize the mouse with isoflurane and disconnect the microscope from the baseplate.
-
97
Coil the cable of the microscope gently to avoid kinking or twisting the wire. Kinks or twists in the wire can result in dropped frames and therefore a loss of data.
-
98
Attach the dust cap to the baseplate, as described in Step 75, and allow the animal to wake up from anesthesia in a secure cage.
Data preprocessing steps and conversion to ΔF/F format
Critical Step
The ideal method for signal extraction and data analysis will depend on multiple variables related to the quality of the data, such as signal-to-noise ratio, general physiological properties of the neurons, such as firing rate, and the anatomical organization of the neurons within the imaging region, such as cell density. Nonetheless, for most data sets, some general pre-processing steps can be applied, as described in Steps 99 and 100 (Supplementary Fig. 10).
-
99
To increase data processing speed, reduce the size of the data files by binning the data in the spatial (i.e., average in space) and/or temporal (i.e., average in time) domain26,29,42 with the use of custom software written in MATLAB29, Mosaic analysis software26 or ImageJ. Spatially binning imaging data can facilitate the analysis process in two ways. First, spatially binning the data can speed up the analysis process by reducing the file size of the movie. Second, spatially binning the data can smooth the edges between contrasting structures, which acts to improve the accuracy of the motion-correction algorithm, especially when a larger number of frameshifts occurs. We have used Mosaic analysis software to spatially bin Ca2+ imaging data collected under imaging conditions that vary by imaging depth, lens diameter, brain region and neuron population, and a spatial bin of 4 works well for most data sets26,29.
Critical Step
Spatial binning of the data will reduce the number of pixels in each frame. Thus, when spatially binning the data, the spatially binned data set should be inspected to ensure that distinct biological structures can still be clearly visualized (Supplementary Fig. 10a). If the sampling rate of the movie is at a higher temporal resolution than is necessary for the analysis of cellular activity, the data can be temporally binned for faster execution of future data processing steps (Supplementary Fig. 10b). For most neuron populations, we acquire data at 15 Hz, and we have found that a temporal bin of 3 (downsample to 5 Hz) sufficiently reduces the size of the data without obscuring transient Ca2+ signals26. Other groups have reported similar downsampling procedures in brain regions such as the hippocampus29.
Critical Step
Temporal binning of the data will reduce the total number of frames and therefore the number of time points in which the signal was sampled. When temporally binning the data, the dynamic properties of the neuronal population, as well as the Ca2+ indicator, should be considered so that the frame rate is sufficient to adequately detect Ca2+ transients. It is not recommended that the data be temporally binned to a rate that is slower than the binding kinetics of the Ca2+ indicator (rise time + half the decay time).
-
100
It is common for drift or frame shifts to occur within a given imaging session, interfering with the detection of biological signals across time. To increase the accuracy of signal detection, reduce motion artifacts through the application of a motion-correction algorithm to either the raw or binned data set63,82. ImageJ and Mosaic analysis software contain plugins or built-in algorithms, respectively, for the motion correction of Ca2+ imaging data26. There are multiple types of motion-correction algorithms that can be used with the above software, such as intensity- or feature-based algorithms that are suitable for Ca2+ imaging data. For most data sets, we have successfully reduced motion artifacts by the application of a rigid body translation algorithm (similar to the TurboReg plugin for ImageJ) available in the Mosaic analysis software package (Supplementary Fig. 10c).
-
101
After completing the pre-processing steps (dimension reduction and motion correction), convert video files to a ΔF/F format26,29,55, which represents the percent change in fluorescence over baseline. This conversion can be accomplished with custom code written in MATLAB29, ImageJ74 or Mosaic analysis software26. See Supplementary Videos 2,3,4,5,6,7 for an example of a raw Ca2+ imaging data set (Supplementary Video 2) that was processed as described in Steps 99 and 100 (Supplementary Videos 3,4,5,6,7,8,9,10). Supplementary Figure 11 provides information that was used to apply the motion-correction algorithm. (See ANTICIPATED RESULTS for more analysis information.)
Anatomical verification of microendoscope placement
-
102
After the completion of an experiment (i.e., all planned imaging sessions within a subject), confirm anatomical placement of the lens above the neuronal population of interest42.
Troubleshooting
Troubleshooting advice can be found in Table 3.
Timing
Steps 1–6, virus dilution study: 3 weeks to 2 months
Steps 7–46, virus injection and lens implantation: 2–3 h
Steps 47–75, attachment of baseplate: 2 h
Steps 76–98, in vivo imaging in freely behaving mice: 1–2 h per session, depending on behavioral paradigm
Steps 99–101, pre-processing steps for data analysis: ∼2 h
Step 102, anatomical verification of microendoscope placement: ∼2 h
Anticipated results
Successful completion of this protocol permits the visualization of active neural ensembles located within both superficial and deep brain regions of freely behaving mice26. Figure 16 shows representative examples of in vivo Ca2+ imaging data collected from cortical (Fig. 16a–f), subcortical (Fig. 16g–i) and deep brain areas (Fig. 16m–r) of mice, as they actively engage with their environment. These data were collected using the procedural steps outlined in this protocol by various members of our laboratory, demonstrating that this protocol can be easily adapted by multiple users and applied to a wide variety of brain structures. Further examples of typical results obtained by our laboratory can be found in reference26. To obtain similar results, in the following section, we briefly describe currently available methods to extract Ca2+ transients from spatially defined cells. We also provide references (when available) to direct readers to studies that have used these data extraction methods in conjunction with imaging procedures similar to those described here (i.e., a miniaturized epifluorescence microscope used in conjunction with a microendoscopic lens).
(a–r) The protocol described here can be adapted for successful in vivo recordings of Ca2+ transients within superficial (a–f), subcortical (g–i) and (j–r) deep brain regions of freely behaving mice. All examples are from one 10-min acute recording session (one mouse per brain region, data acquired at 15 Hz, temporally down sampled to 5 Hz, spatial bin of 4). A 1-mm-diameter lens was used to image Ca2+ signals in superficial and subcortical areas, whereas a 0.5-mm-diameter lens was used in deep brain regions. The left column shows digital images generated by creating a mean intensity projection of fluorescence activity across the entire recording session, the middle column shows the spatial location of optically detected GCaMP expressing cells, and the right column shows activity traces (variation from the mean fluorescence signal over time) of cells outlined in the corresponding spatial map. Spatial maps and activity traces were extracted with PCA/ICA analysis (Mosaic analysis software, Inscopix). (a–c) Somatosensory cortex neurons were targeted by injecting AAVdj-CamkII-GCaMP6f (titer: 5.6 × 1012, 1:8 dilution, UNC vector core) into the barrel field region of an adult wild-type (WT) male mouse. (d–f) Prefrontal cortex neurons were targeted by injecting AAV-DJ-CamkIIa-GCaMP6s into the prefrontal cortex of an adult WT male mouse (titer: 5.3 × 1012, 1:6 dilution, UNC vector core). (g–i) AAV-DJ-CamkIIa-GCaMP6m was injected into the dentate gyrus of a WT male mouse (titer: 7.0 × 1012, 1:1 dilution, Stanford). (j–l) GCaMP6m was targeted to GABAergic neurons within the lateral hypothalamus (LH) by injecting AAVdj-EF1a-DIO-DIO-GCaMP6m (titer 9.7 × 1012, 1:2 dilution, Stanford) into the LH of an adult VGAT-Cre male mouse. (m–o) Dopamine neurons within the ventral tegmental area (VTA) of an adult male mouse were targeted by injecting AAVdj-EF1a-DIO-DIO-GCaMP6m (titer 9.7 × 1012, 1:2 dilution, Stanford) into the VTA of a TH-Cre mouse. (p–r) Bed nucleus of the stria terminalis (BNST) neurons were targeted by injecting AAV-DJ-CamkIIa-GCaMP6s (titer: 5.3 × 1012, 1:6 dilution, UNC vector core) into the BNST of an adult WT male mouse. Notice that deep brain regions differ from cortical regions in image clarity. Injection volumes for all regions were 300 μl. Scale bars, 125 μm. All procedures were approved by UNC IACUC.
Extraction of individual cellular Ca2+ signals from high-dimension data sets
An advantage of Ca2+ imaging is the potential to simultaneously monitor the activity of large neural networks. However, this sampling method produces data sets of high dimensionality (npixels × nframes), and extraction of meaningful biological signals from imaging data requires the deconvolution of the data into independent signal sources83,84. This task can be difficult for multiple reasons. First, single-photon microscopy lacks optical sectioning abilities14,20,23, resulting in the simultaneous detection of fluorescence signals from cells located within and outside the focal by the photosensor located within the head-mounted microscope83 (Supplementary Fig. 12a). This aspect of one-photon microscopy increases the potential for signal cross talk, and methods for disentangling the neural dynamics of optically detected cells within the focal plane from those out of the focal plane are therefore necessary. Cellular cross talk can also occur if cells within the focal plane overlap in anatomical space, as these cells will have overlapping pixel sets83. This specific potential for signal cross talk is particularly high when imaging densely populated networks (i.e., >50 cells in the field of view). Finally, the large size of Ca2+ imaging data sets (many pixels collected over multiple frames) can make the analysis process extremely labor-intensive (Supplementary Fig. 12b). Thus, to facilitate the extraction of transient Ca2+ activity from spatially defined biological structures, multiple data analysis methods and computational techniques have been developed83,84,85,86 (Supplementary Fig. 12c). Below, we briefly describe some of the currently available methods for decomposing high-dimensionality imaging data into individual cellular signals.
A conventional method for segmenting Ca2+ imaging data into individual cellular signals is region-of-interest (ROI) analysis. With ROI analysis, spatial clusters of pixels are assigned to individual cells, by visually inspecting the data and by manually circling pixels that have characteristic morphological properties of a GCaMP-expressing cell. Temporal information related to dynamic Ca2+ activity is then extracted by converting pixel intensity values within an identified ROI into a ΔF/F trace32. This analysis method can be conducted with Mosaic analysis software, which is a free plugin available for ImageJ74, or custom software written in MATLAB or Python55. Importantly, ROI analysis has been used to successfully extract transient Ca2+ activity from individual cells that were visualized with imaging procedures similar to those described in this protocol. Specifically, it was used to extract the activity of individual neurons located within the entorhinal cortex of male mice (∼40 per mouse)74, as well as the activity pattern of neurons located within the premotor nucleus of male zebra finches (reported 167 neurons across 9 birds; estimated to be ∼18 per bird)55.
Although ROI analysis has been successfully used to isolate individual cellular Ca2+ transients from data sets that contain a relatively low number of identified cells (<40 per subject), it may not be well suited or feasible to extract individual cellular signals from Ca2+ imaging data sets in which the activity of a large number of independent cellular signals can be detected. Specifically, for Ca2+ imaging with data sets that contain a high density of GCaMP-expressing cells26,29, ROI analysis may not permit cells that have full or partially overlapping boundaries with each other to be manually disentangled. In addition, because of a lack of optical sectioning, it is possible for fluctuations in fluorescence intensity within an ROI to be associated with a cell that was manually identified by the experimenter, as well as fluctuations in neural activity that are occurring above and/or below this cell. To account for these limitations in ROI analysis, efforts have been made to reduce signal cross talk with ROI analysis through semiautomated morphological filtering of cellular signals85: the use of temporal cross-correlation algorithm to identify highly correlated pixels within an ROI86 and computational methods to correct for signal contamination by out-of-focus fluorescence87. Finally, because the user must manually define the spatial location of each cell, this type of analysis can be extremely labor-intensive83. Thus, ROI analysis is best suited for data sets that contain relatively sparse populations of neurons that do not overlap in space55.
For large data sets that contain a high density of neurons, automated cell-sorting algorithms may be better suited to extract spatial and temporal properties of individual cellular Ca2+ signals83,88. For example, two software packages, Thunder52 (see Supplementary Methods for code) and Mosaic (Inscopix), provide automated cell-sorting algorithm to extract data from digitally collected images. In brief, this algorithm first runs principal component analysis (PCA) to reduce the dimensionality of the data by removing signals that mostly encode noise. Dimension reduction is then followed by independent component analysis (ICA) to identify the spatial and temporal properties of visualized cells by segmenting the data into statistically independent spatial and temporal signals83. The extracted signals, or independent components, are then manually sorted to confirm that the extracted data are representative of cellular Ca2+ transients from spatially defined GCaMP-expressing cells26,29. Because PCA/ICA analysis relies on statistical independence of signals, it outperforms ROI analysis in the reduction of signal cross talk, and it can be used to relatively quickly extract independent cellular signals from relatively large data sets83. For example, for optical imaging data collected with the methods described in this protocol, PCA/ICA analysis has been used to successfully extract signals from up to 1,000 neurons in the hippocampus29,42, hundreds of neurons in the lateral hypothalamus26 and hundreds of neurons in the medial entorhinal cortex64. In addition to these densely populated networks, PCA/ICA analysis has also been used to successfully extract individual cellular signals from relatively sparse neuronal populations, such as AGRP neurons located within the arcuate nucleus of the hypothalamus (61–99 neurons in four mice)27.
Although PCA/ICA is superior to ROI analysis in some areas of data extraction (i.e., analysis speed and cross-talk reduction), it is important to note that this computational method also has limitations that can reduce the accuracy of data interpretation. First, the number of independent components to be extracted from the data set must be specified by the experimenter before the analysis. This is usually done by estimating the number of active cells after visual inspection of the digitally acquired data set and setting the number of independent components to be larger than this number26,29,64. As can be expected, the quality of cell extraction is partially dependent on this estimate. For instance, if the number of independent components is set to be much larger than the number of cells in the data, PCA/ICA may separate individual cells into multiple components. In addition, the PCA/ICA algorithm is stochastic. Hence, there is a high probability that the extracted components will not exactly be the same across multiple runs of the algorithm, even on the same data set. Thus, when applying this algorithm to data sets collected across multiple imaging sessions, a conservative approach may be to first run PCA/ICA to extract the cellular signals and then, on the basis of the cells' spatial filter (i.e., x,y pixel coordinates associated with that cell), only select cells that have been consistently identified across imaging sessions27. Finally, because this analysis method relies on the identification of statistically independent signals, it may not be well suited for the analysis of neural ensembles with low activity levels that do not substantially differ from baseline83. In this case, ROI analysis may be better suited to extract Ca2+ activity traces from independent cellular signals.
Other published automated cell-sorting algorithms include non-negative matrix factorization84 or a matrix factorization that explicitly models Ca2+ fluorescence dynamics88. As yet, no paper has systematically benchmarked these methods against each other. Hence, it is currently unclear which method is best suited for Ca2+ indicator data from epifluorescence microendoscopy. In addition, to our knowledge, this analysis method has also not been used to analyze Ca2+ imaging data obtained by the methods described in this protocol.
Correlation of neural activity with experimental manipulations, environmental context or behavioral events
Once the spatial and temporal data have been extracted, the activity of cells can be correlated with contextual changes in environment64,74, environmental exploration26,42,64,75, time-locked stimulus presentations or behavioral events26, learning- or experience-mediated plasticity within networks29,55, or pharmacological induced alterations of network activity42 (Fig. 17a). Perhaps the most notable advantage provided by this technique is the ability to further analyze the data in relation to the spatial location of cells, thus providing activity maps or information related to functional connectivity within identified microcircuits (Fig. 17b; see Supplementary Data 1 for GUI to conduct such analysis and Supplementary Methods for description of GUI contents and instructions). Importantly, the ability to image cell type–specific activity during naturalistic animal behavior allows this imaging protocol to be applied to a wide array of experimental manipulations in order to answer questions that could not be explored with previous methods for monitoring of neural activity.
The methods outlined in this procedure permit the activity of neurons to be monitored with cellular-level resolution as mice perform naturalistic behaviors. (a) When this method is further combined with simultaneous time-locked recordings of environmental, behavioral or pharmacological events, the activity of optically detected neurons can be sorted by their response profile to specific stimuli or behavioral events. In this example, GABAergic neurons within the lateral hypothalamus of adult male mice (n = 6) were monitored during consummatory licks for a sucrose reward. 75 cells out of 743 total identified cells showed excitatory responses to reward consumption. The average response of each cell was averaged across trials and aligned to the lick response (5 s before and 10 s after). (b) Cell activity map from a representative animal shows response of each neuron to consumption of sucrose reward (average difference in fluorescence intensity 1.5 s before and after the event). All procedures were approved by UNC IACUC. See Supplementary Methods for instructions on how to conduct similar analysis. Panels a,b are adapted with permission from Jennings et al.26.
References
Chorev, E., Epsztein, J., Houweling, A.R., Lee, A.K. & Brecht, M. Electrophysiological recordings from behaving animals–going beyond spikes. Curr. Opin. Neurobiol. 19, 513–519 (2009).
Hubel, D.H. Single unit activity in lateral geniculate body and optic tract of unrestrained cats. J. Physiol. 150, 91–104 (1960).
Clark, J.J. et al. Chronic microsensors for longitudinal, subsecond dopamine detection in behaving animals. Nat. Methods 7, 126–129 (2010).
Robinson, D.L., Venton, B.J., Heien, M.L.A.V. & Wightman, R.M. Detecting subsecond dopamine release with fast-scan cyclic voltammetry in vivo. Clin. Chem. 49, 1763–1773 (2003).
Miyawaki, A. et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887 (1997).
Akerboom, J. et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 32, 13819–13840 (2012).
Akemann, W., Mutoh, H., Perron, A., Rossier, J. & Knöpfel, T. Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins. Nat. Methods 7, 643–649 (2010).
Woolum, J.C. & Strumwasser, F. Membrane-potential-sensitive dyes for optical monitoring of activity in Aplysia neurons. J. Neurobiol. 9, 185–193 (1978).
Ohkura, M. et al. Genetically encoded green fluorescent Ca2+ indicators with improved detectability for neuronal Ca2+ signals. PLoS ONE 7, e51286 (2012).
Tian, L. et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 6, 875–881 (2009).
Chen, J.L., Andermann, M.L., Keck, T., Xu, N.-L. & Ziv, Y. Imaging neuronal populations in behaving rodents: paradigms for studying neural circuits underlying behavior in the mammalian cortex. J. Neurosci. 33, 17631–17640 (2013).
Podor, B. et al. Comparison of genetically encoded calcium indicators for monitoring action potentials in mammalian brain by two-photon excitation fluorescence microscopy. Neurophotonics 2, 021014 (2015).
Chen, T.-W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Grienberger, C. & Konnerth, A. Imaging calcium in neurons. Neuron 73, 862–885 (2012).
Palmer, A.E. & Tsien, R.Y. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat. Protoc. 1, 1057–1065 (2006).
Cichon, J. & Gan, W.-B. Branch-specific dendritic Ca2+ spikes cause persistent synaptic plasticity. Nature 520, 180–185 (2015).
Smetters, D., Majewska, A. & Yuste, R. Detecting action potentials in neuronal populations with calcium imaging. Methods 18, 215–221 (1999).
Ziv, Y. & Ghosh, K.K. Miniature microscopes for large-scale imaging of neuronal activity in freely behaving rodents. Curr. Opin. Neurobiol. 32, 141–147 (2015).
Hamel, E.J.O., Grewe, B.F., Parker, J.G. & Schnitzer, M.J. Cellular level brain imaging in behaving mammals: an engineering approach. Neuron 86, 140–159 (2015).
Jung, J.C., Mehta, A.D., Aksay, E., Stepnoski, R. & Schnitzer, M.J. In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy. J. Neurophysiol. 92, 3121–3133 (2004).
Levene, M.J., Dombeck, D.A., Kasischke, K.A., Molloy, R.P. & Webb, W.W. In vivo multiphoton microscopy of deep brain tissue. J. Neurophysiol. 91, 1908–1912 (2004).
Resendez, S.L. & Stuber, G.D. In vivo calcium imaging to illuminate neurocircuit activity dynamics underlying naturalistic behavior. Neuropsychopharmacology 40, 238–239 (2015).
Barretto, R.P.J. & Schnitzer, M.J. In vivo optical microendoscopy for imaging cells lying deep within live tissue. Cold Spring Harb. Protoc. 2012, 1029–1034 (2012).
Jung, J.C. & Schnitzer, M.J. Multiphoton endoscopy. Opt. Lett. 28, 902–904 (2003).
Jennings, J.H. & Stuber, G.D. Tools for resolving functional activity and connectivity within intact neural circuits. Curr. Biol. 24, R41–50 (2014).
Jennings, J.H. et al. Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell 160, 516–527 (2015).
Betley, J.N. et al. Neurons for hunger and thirst transmit a negative-valence teaching signal. Nature 521, 180–185 (2015).
Ghosh, K.K. et al. Miniaturized integration of a fluorescence microscope. Nat. Methods 8, 871–878 (2011).
Ziv, Y. et al. Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci. 16, 264–266 (2013).
Svoboda, K. & Yasuda, R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron 50, 823–839 (2006).
Denk, W. et al. Anatomical and functional imaging of neurons using 2-photon laser scanning microscopy. J. Neurosci. Methods 54, 151–162 (1994).
Dombeck, D.A., Khabbaz, A.N., Collman, F., Adelman, T.L. & Tank, D.W. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron 56, 43–57 (2007).
Dombeck, D. & Tank, D. Two-photon imaging of neural activity in awake mobile mice. Cold Spring Harb. Protoc. 2014, 726–736 (2014).
Cui, G. et al. Deep brain optical measurements of cell type-specific neural activity in behaving mice. Nat. Protoc. 9, 1213–1228 (2014).
Gunaydin, L.A. et al. Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551 (2014).
Buzsáki, G. et al. Tools for probing local circuits: high-density silicon probes combined with optogenetics. Neuron 86, 92–105 (2015).
Cunningham, J.P. Analyzing neural data at huge scale. Nat. Methods 11, 911–912 (2014).
Helmchen, F., Borst, J.G. & Sakmann, B. Calcium dynamics associated with a single action potential in a CNS presynaptic terminal. Biophys. J. 72, 1458–1471 (1997).
Sparta, D.R. et al. Construction of implantable optical fibers for long-term optogenetic manipulation of neural circuits. Nat. Protoc. 7, 12–23 (2012).
Jennings, J.H. et al. Distinct extended amygdala circuits for divergent motivational states. Nature 496, 224–228 (2013).
Stuber, G.D. & Mason, A.O. Integrating optogenetic and pharmacological approaches to study neural circuit function: current applications and future directions. Pharmacol. Rev. 65, 156–170 (2013).
Berdyyeva, T. et al. Zolpidem reduces hippocampal neuronal activity in freely behaving mice: a large-scale calcium imaging study with miniaturized fluorescence microscope. PLoS ONE 9, e112068 (2014).
Alivisatos, A.P. et al. The brain activity map project and the challenge of functional connectomics. Neuron 74, 970–974 (2012).
Barretto, R.P.J. et al. Time-lapse imaging of disease progression in deep brain areas using fluorescence microendoscopy. Nat. Med. 17, 223–228 (2011).
Roth, G. & Dicke, U. Evolution of the brain and intelligence. Trends Cogn. Sci. 9, 250–257 (2005).
Mank, M. et al. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat. Methods 5, 805–811 (2008).
Vorhees, C.V. & Williams, M.T. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat. Protoc. 1, 848–858 (2006).
Stuber, G.D. Dissecting the neural circuitry of addiction and psychiatric disease with optogenetics. Neuropsychopharmacology 35, 341–342 (2010).
McCall, J.G. et al. Fabrication and application of flexible, multimodal light-emitting devices for wireless optogenetics. Nat. Protoc. 8, 2413–2428 (2013).
Leung, L.C., Wang, G.X. & Mourrain, P. Imaging zebrafish neural circuitry from whole brain to synapse. Front. Neural Circuits 7, 7 (2013).
Ferguson, A.R., Nielson, J.L., Cragin, M.H., Bandrowski, A.E. & Martone, M.E. Big data from small data: data-sharing in the 'long tail' of neuroscience. Nat. Neurosci. 17, 1442–1447 (2014).
Freeman, J. et al. Mapping brain activity at scale with cluster computing. Nat. Methods 11, 941–950 (2014).
Sejnowski, T.J., Churchland, P.S. & Movshon, J.A. Putting big data to good use in neuroscience. Nat. Neurosci. 17, 1440–1441 (2014).
Müller, J., Bakkum, D.J. & Hierlemann, A. Sub-millisecond closed-loop feedback stimulation between arbitrary sets of individual neurons. Front. Neural Circuits 6, 121 (2013).
Markowitz, J.E. et al. Mesoscopic patterns of neural activity support songbird cortical sequences. PLoS Biol. 13, e1002158 (2015).
Wang, C. & Ji, N. Characterization and improvement of three-dimensional imaging performance of GRIN-lens-based two-photon fluorescence endomicroscopes with adaptive optics. Opt. Express 21, 27142 (2013).
Zariwala, H.A. et al. A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J. Neurosci. 32, 3131–3141 (2012).
Madisen, L. et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 15, 793–802 (2012).
Madisen, L. et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015).
Chen, Q. et al. Imaging neural activity using Thy1-GCaMP transgenic mice. Neuron 76, 297–308 (2012).
Gee, J.M. et al. Imaging activity in astrocytes and neurons with genetically encoded calcium indicators following in utero electroporation. Front. Mol. Neurosci. 8, 10 (2015).
Witten, I.B. et al. Recombinase-driver rat lines: tools, techniques, and optogenetic application to dopamine-mediated reinforcement. Neuron 72, 721–733 (2011).
Thévenaz, P., Ruttimann, U.E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (1998).
Sun, C. et al. Distinct speed dependence of entorhinal island and ocean cells, including respective grid cells. Proc. Natl. Acad. Sci. USA 112, 9466–9471 (2015).
Ewald, A.J., Werb, Z. & Egeblad, M. Monitoring of vital signs for long-term survival of mice under anesthesia. Cold Spring Harb. Protoc. 2011 doi:10.1101/pdb.prot5563 (1 February 2011).
Wojtowicz, J.M. & Kee, N. BrdU assay for neurogenesis in rodents. Nat. Protoc. 1, 1399–1405 (2006).
Li, Y. et al. Direct labeling and visualization of blood vessels with lipophilic carbocyanine dye DiI. Nat. Protoc. 3, 1703–1708 (2008).
Burger, C. et al. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol. Ther. 10, 302–317 (2004).
Royo, N.C. et al. Specific AAV serotypes stably transduce primary hippocampal and cortical cultures with high efficiency and low toxicity. Brain Res. 1190, 15–22 (2008).
Malik, S.Z., Maronski, M.A., Dichter, M.A. & Watson, D.J. in Neurotrophic Factors (ed. Skaper, S.D.) 846, 305–319 (Humana Press, 2012).
Athos, J. & Storm, D.R. High-precision stereotaxic surgery in mice. Curr. Protoc. Neurosci. 4, A.4A.1–A.4A.9 (2001).
Holtmaat, A. et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat. Protoc. 4, 1128–1144 (2009).
Paxinos, G., Franklin, K.B.J. & Franklin, K.B.J. The Mouse Brain in Stereotaxic Coordinates (Academic Press, 2001).
Kitamura, T. et al. Entorhinal cortical ocean cells encode specific contexts and drive context-specific fear memory. Neuron 87, 1317–1331 (2015).
Andermann, M.L. et al. Chronic cellular imaging of entire cortical columns in awake mice using microprisms. Neuron 80, 900–913 (2013).
Chia, T.H. & Levene, M.J. Microprisms for in vivo multilayer cortical imaging. J. Neurophysiol. 102, 1310–1314 (2009).
Chia, T.H. & Levene, M.J. In vivo imaging of deep cortical layers using a microprism. J. Vis. Exp. doi:10.3791/1509 (27 August 2009).
Pacary, E. et al. Visualization and genetic manipulation of dendrites and spines in the mouse cerebral cortex and hippocampus using in utero electroporation. J. Vis. Exp. 65, e4163 doi:10.3791/4163 (26 July 2012).
Schneider Gasser, E.M. et al. Immunofluorescence in brain sections: simultaneous detection of presynaptic and postsynaptic proteins in identified neurons. Nat. Protoc. 1, 1887–1897 (2006).
Kim, T.I. et al. Injectable, cellular-scale optoelectronics with applications for wireless optogenetics. Science 340, 211–216 (2013).
Fenno, L., Yizhar, O. & Deisseroth, K. The development and application of optogenetics. Annu. Rev. Neurosci. 34, 389–412 (2011).
Guizar-Sicairos, M., Thurman, S.T. & Fienup, J.R. Efficient subpixel image registration algorithms. Opt. Lett. 33, 156–158 (2008).
Mukamel, E.A., Nimmerjahn, A. & Schnitzer, M.J. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63, 747–760 (2009).
Maruyama, R. et al. Detecting cells using non-negative matrix factorization on calcium imaging data. Neural Netw. 55, 11–19 (2014).
Ohki, K., Chung, S., Ch'ng, Y.H., Kara, P. & Reid, R.C. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature 433, 597–603 (2005).
Ozden, I., Lee, H.M., Sullivan, M.R. & Wang, S.S.-H. Identification and clustering of event patterns from in vivo multiphoton optical recordings of neuronal ensembles. J. Neurophysiol. 100, 495–503 (2008).
Pinto, L. & Dan, Y. Cell-type-specific activity in prefrontal cortex during goal-directed behavior. Neuron 87, 437–450 (2015).
Pnevmatikakis, E. et al. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89, 285–299 (2016).
Acknowledgements
We acknowledge J. Horwitz for his assistance with figure preparation; L. Eckman and S. Pfarr for their assistance with data collection; and L. Cody, S. Otte, A. Stamatakis, A. Crowther, J. Parker, I. Witten, J. Freeman, V. Ghukasyan, R. van Zessen, A. Hamid, C. Vanderwheel and members of the Stuber Lab for helpful discussion. This work was supported by The Klarman Family Foundation, The Brain and Behavior Research Foundation, The Simon's Foundation, The Foundation of Hope, the National Institute on Drug Abuse (DA032750 and DA038168), the National Institute of Child Health (NICHD) and Human Development (HD079124) and the National Institute on Alcohol Abuse and Alcoholism (P60 AA011605). J.H.J. was supported by National Institute of Mental Health (NIMH) grant MH104013. S.L.R. was supported by NICHD grant T32 HD040127, Postdoctoral Research in Neurodevelopmental Disorders.
Author information
Authors and Affiliations
Contributions
S.L.R. and G.D.S. wrote the manuscript. S.L.R., J.H.J., R.L.U. and V.M.K.N. contributed to the development of the procedure. Z.C.Z., J.M.O., H.N., J.A.M. and O.K. assisted with manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
G.D.S. is the recipient of a DECODE grant from Inscopix, Palo Alto, CA.
Integrated supplementary information
Supplementary Figure 1 Preparation of surgery tools for lens handling and bone removal.
(a) Digital image depicting the tips of a pair of forceps that have been covered with ~10 mm of heat shrink. (b) Bulldog serrefine covered with ~5 mm of heat shrink. (c) Heat shrink covered bulldog serrefine tightly clamped in guide cannula holder. (d) Guide cannula holder with secured bulldog serrefine attached to a stereotaxic apparatus. (e) 27G needle bent at a 45° angle. Prepare needle before surgery and use to scoop out bone fragments after and peel away dura.
Supplementary Figure 2 Confirmation of cell type specific expression of GCaMP.
(a) Confocal image stack of GCaMP expression within the VTA of a TH-Cre adult male mouse. (b) Confocal image stack of VTA neurons labeled with GCaMP6m (left) and stained for TH (center). Merged image shows co-labeling of GCaMP6m and TH indicating that GCaMP6m was successfully expressed within VTA-DA neurons (AAVdj-Ef1a-GCaMP6m; 1:2 dilution, 4.8 x 1013). All procedures were approved by UNC IACUC.
Supplementary Figure 3 Visualization of Ca2+ transients through either a 0.5 or 1 mm lens implanted in the prefrontal cortex.
Fluorescently encoded Ca2+ dynamics were monitored over a 10-min acute recording session (15 Hz, down sampled to 5Hz) in adult wild type males. Either a (a) 0.5 or (b) 1 mm diameter lens was used to visualize fluorescently encoded Ca2+ signals (AAV-DJ-CamkIIa-GCaMP6s, 5.3 x 1012, 1:4 dilution, UNC vector core). Top row shows mean intensity projections of fluorescence signals across the entire 10-min session (spatial bin of 4). Notice the difference in contrast between anatomical structures that is conveyed by each of the lenses. PCA/ICA analysis was used to extract the spatial location (middle row) and activity traces (bottom row) of individual cells. 1.0 mm diameter lenses (right) have larger fields of view than 0.5 mm diameter lenses (left) and therefore have the potential to detect a larger number of cells. However, the quality of spatial and temporal filters is similar between lenses. Optical distortion of cellular signals along the edge of the lens occurs with both 0.5 and 1 mm diameter lenses. All procedures were approved by UNC IACUC.
Supplementary Figure 4 Position of injector needle relative to lens placement.
(a) To reduce damage under the lens, it is recommended that viral injections are not made directly under the lens. Rather, injections should be placed 250 μm lateral and ventral to the placement of the lens.
Supplementary Figure 5 Visualization of brain tissue and vasculature during lens implant.
(a) A 1 mm diameter lens was implanted in the prefrontal cortex of an adult wild type male. Throughout the duration of the lens implant procedure, tissue and vasculature should be visible through the lens. These can be visualized by looking directly through the top of the lens with the naked eye or visualized by centering the surgical microscope above the top of the lens and looking through the microscope objectives. All procedures were approved by UNC IACUC.
Supplementary Figure 6 Cell death following long term expression of GCaMP in the somatosensory cortex.
In vivo imaging of GCaMP encoded Ca2+ dynamics (AAVdj-CamkII-GCaMP6f, Titer: 5.6 x 1012, 1:4 dilution, UNC vector core) was conducted over a 10-min acute recording session (15 Hz, down sampled to 5Hz) 2 (top row) and 12 (bottom row) months following virus injection and lens implantation surgery. (a) Digital images of mean intensity pixel projections across the entire 10-min session (spatial bin of 4). Following 12 months of GCaMP expression (bottom), distinct biological structures are no longer visible. (b,c) PCA/ICA analysis was used to extract the spatial location and activity traces of individual cells (Mosaic Analysis Software, Inscopix). Because spatial or temporal characteristics of GCaMP expressing cells were not identifiable following 12 months of viral expression, independent components (ICs) extracted from this data set could not be sorted by these features. Thus, to compare ICs extracted at each time point, the first 125 ICs were selected from each data set. (b) Merged image of spatial filters for all extracted ICs 2 and 12 months following surgery. At 2 months (top), spatial filters of extracted ICs have morphological characteristics of cells. In contrast, defined anatomical structures are not visible in this same mouse 12 months following surgery (bottom). (c) Similarly, representative activity traces show temporal dynamics characteristic of Ca2+ signals at 2, but not at 12 months following surgery. All procedures were approved by UNC IACUC.
Supplementary Figure 7 Practice baseplate attachment surgery.
(a) Secure the dummy microscope in the microscope gripper connected to a micromanipulator. (b) Prepare a glass slide with a dummy head cap. Position the dummy microscope over a glass slide with the microscope placed about the same distance that would be necessary to appropriately attach a baseplate to the head cap of a mouse (~2 mm above the dummy head cap). (c) Attach the dummy microscope to the glass slide in a similar manner that would be used in an actual baseplate surgery.
Supplementary Figure 8 Habituation to microscope attachment.
(a) Affix a wire cable to the dummy microscope with super glue. (b) Anesthetize mouse and attach dummy microscope to baseplate. When mouse is ambulatory, place the mouse in the test chamber and tether the wire cable to the ceiling. Continue habituation procedures until the mouse can move naturally through the environment while tethered to the dummy microscope.
Supplementary Figure 9 Repeated imaging of GCaMP encoded Ca2+ signals in the somatosensory cortex.
In vivo imaging of GCaMP encoded Ca2+ dynamics (AAVdj-CamkII-GCaMP6f, titer: 5.6 x 1012, 1:4 dilution, UNC vector core) within the somatosensory cortex of an adult wild type male was monitored over 10-min recording sessions (15 Hz, down sampled to 5Hz). One recording was made per day over a 5 day period. (a) Mean intensity projections across the entire 10-min session. Similar anatomical landmarks could be repeatedly visualized in the imaging data. (b) The spatial location of GCaMP expressing cells was extracted with PCA/ICA analysis (Mosaic Analysis Software, Inscopix). On each day, cells located within similar anatomical space could be visualized. However, the number of cells extracted with PCA/ICA analysis varied across sessions. (c) The extracted spatial maps were saved as individual TIFF files and converted to a TIFF stack in ImageJ. TurboReg motion correction algorithm (ImageJ) was applied to the maps for alignment. (d) Example of spatial maps from recording day 2 (red) and (5) merged on top of each other. Prior to motion correction, many similarly detected cells are misaligned. (e) Application of the TurboReg motion correction algorithm increases the number of aligned cells.
Supplementary Figure 10 Pre-processing of digitally acquired Ca2+ imaging data.
While multiple data analysis methods exist to extract independent cellular components, there are general data pre-processing steps that can be applied to in vivo Ca2+ imaging data sets to facilitate future data analysis steps as well as increase the accuracy of data interpretation. These steps include (a,b) down sampling of the data to increase the speed of data processing as well as (b) the application of a motion correction algorithm, which increases signal detection accuracy. (a) If images contain a greater number of pixels than is required to visualize distinct biological structures, such as cells and vasculature, the data can be spatially binned to reduce the file size and facilitate future data analysis steps. We have found that a spatial bin of 4 works well for most neuron populations. Notice that spatial binning of the data by a factor of 4 does not interfere with the detection of distinct biological structures. (b) If more data samples were acquired than is necessary for the detection of GCaMP encoded Ca2+ transients, the data can be further down sampled in the temporal domain. For data acquired at 15 Hz, we have found that a temporal bin of 3 (down sample to 5 Hz) adequately reduces the file size while still allowing the detection of individual Ca2+ transients. (c) It is common for small frame shifts or drift in the focal plane to occur across a recording session. To realign images, and therefore the location of biological structures, a motion correction algorithm can be applied to the data set. The data shown in this paper was aligned via rigid body translation (Mosaic Analysis Software). In brief, this algorithm works by aligning all frames to a reference region (outlined in blue) that was selected from a reference frame in the data set. All frames in the data set (target images) are translated in the x,y plane to align each target image to the reference image on the basis of pixel intensity distribution within the reference region. All procedures were approved by UNC IACUC.
Supplementary Figure 11 Data processing information for supplemental video 4 and 6.
A rigid body motion correction algorithm was applied to the spatially binned Ca2+ imaging data shown in video 4 (Mosaic, Inscopix). (a) Reference image used for motion correction algorithm. (b) Region outlined in blue represents the area of the reference frame that was selected as the reference region. A large blood vessel was selected as its spatial location was stable throughout the imaging session and it exhibits a high contrast in pixel intensity compared to the surrounding neural tissue. Note – to facilitate selection of the reference region, the contrast was enhanced and the pixel intensity was inverted by multiplying all pixels by a value of -1. (c) Distance in pixels that each frame was translated in the x plane. (d) Distance in pixels that each frame was translated in the y plane. (e) Mean intensity projection (F’0) used to convert the motion corrected and cropped video (Supplemental video 5) to a ΔF/F format. All procedures were approved by UNC IACUC.
Supplementary Figure 12 Extraction of individual cellular Ca2+ signals from high dimension data sets.
(a) The photosensor (CMOS sensor) is composed of an array of photodetectors (pixels) that encode the spatial location and intensity of all detected emission signals that are captured during a defined exposure period. Photonic data collected during this exposure period are converted to a digital signal and stored as a TIFF file. (b) Each TIFF (left) file contains information regarding how physiological structures under the lens are distributed in anatomical space as well as the intensity of fluorescence emissions from these structures at a defined point in time. The sequential collection of these images (right) enables changes in fluorescence intensity, and therefore neural activity, to be extrapolated from changes in pixel intensity over time. However, this requires methods to extract meaningful biological signals from high dimensionality Ca2+ imaging data sets. (c) Example of two analysis methods for the extraction of independent cellular signals. Briefly, region of interest analysis (left) requires the user to manually identify pixels associated with an individual cell. Pixels within this region of interest are then converted to DF/F activity traces. Analysis method on the left shows on example of an automated cell sorting algorithm (PCA/ICA) which extracts the spatial location and activity traces of individual cells based on statistical independence.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12 Supplementary Methods (PDF 2795 kb)
Visualization of cortical Ca2+ signals in a freely behaving mouse
Fluorescently encoded Ca2+ signals were visualized through a 1 mm diameter GRIN lens implanted in the prefrontal cortex (PFC) of a wild type (WT) male mouse. The recording (nVista Acquisition Software, Inscopix) was made 8 weeks following surgical procedures for the microinjection of virally encoded GCaMP (AAV-DJ-CamkIIa-GCaMP6s, titer: 5.3 × 1012, 1:6 dilution, UNC vector core) and the subsequent implantation of a microendoscopic lens above the PFC. Images were acquired at 15 Hz and processed (spatial bin by 2, motion corrected, cropped, conversion to ΔF/F format) with Mosaic Data Analysis Software (Inscopix). Notice that in many of the active cells, nuclear exclusion of GCaMP (dark region in the center of cell) can be visualized indicating that the monitored cells are healthy. All procedures were approved by UNC IACUC. (MOV 11001 kb)
Unprocessed in vivo Ca2+ imaging movie.
Movie showing an example of raw Ca2+ imaging data collected at 15 Hz (nVista Acquisition Software, Inscopix) from the dentate gyrus of a freely behaving wild-type male mouse (AAV-DJ-CamkIIa-GCaMP6m; titer: 7.0 × 1012, 1:1 dilution, Stanford). Fluorescently encoded Ca2+ signals were visualized through a 1 mm diameter GRIN lens with a miniature, microscope secured to the head of the mouse. Notice that small fluctuations in the spatial location of biological structures occur throughout the movie. All procedures were approved by UNC IACUC. (MOV 8387 kb)
Spatial down sampling of video 2.
Imaging data shown in video 2 was spatially binned (i.e., averaged in space) by a factor of 4 to reduce the file size and increase the rate of subsequent data processing steps (Mosaic Analysis Software, Inscopix). Spatial binning of the movie by a factor of 4 as shown here does not interfere with the ability to detect distinct biological structures, such as cells or vasculature. In addition to increasing data processing speed, spatial binning can also facilitate the motion correction process by smoothing edges (i.e., variations in pixel intensity) between contrasting structures. (MOV 8200 kb)
Motion correction of video 3.
Rigid body translation was used to correct lateral translations (i.e., translations in the x,y plane) within movie frames that may occur as a result of rapid frame shifts during vigorous animal movement or drift in the imaging plane throughout the recording session (Mosaic Analysis Software). The motion correction algorithm used here works by pulling pixels (weighted by intensity) in each movie frame to align with the spatial distribution of pixel intensities within a reference region. To run the algorithm, a reference frame was selected from the movie in which low levels of neural activity were detected and the location of biological structures, such as blood vessels, was representative of their location throughout the majority of the movie. A reference region was then selected from this frame in which all images in the movie were registered to (Supplementary Fig. 11). In general, a large biological structure that is stably detected within all frames as well as exhibits a high contrast in pixel intensity form the surrounding neural tissue, such as a blood vessel, is well suited for the image registration process. Following selection of both the reference frame and region, a motion correction algorithm was applied to register all movie frames. Application of this algorithm resulted in the realignment of biological structures, indicated by the stable spatial location of blood vessels within each frame. The realignment of the target images also resulted in the translation of some frames out of the field of view, indicated by black areas along the edges of the frame that do not encode for digitally collected data. To exclude portions of the image that do not encode for fluorescently detected signals, crop the movie as shown in video 5. (MOV 8075 kb)
Cropped movie after motion correction.
The motion corrected movie shown in video 4 was cropped to exclude portions of the movie that do not encode biologically relevant information (Mosaic Analysis Software). Cropping the movie reduces the number of pixels in each frame, resulting in a smaller file size and enhanced data processing speeds. (MOV 8527 kb)
Conversion of video 5 to ΔF/F format.
To determine the percent change in fluorescence over time, convert the movie file to a ΔF/F format (ΔF = (F'(t) – F'0)/F'0). This is accomplished by first creating a mean projection of pixel intensity across all the frames in the movie (F'0) (Supplementary Fig. 11). Next, for every frame in the movie (F'(t)), subtract the value of the pixel intensity encoded by the mean projection image and subsequently divide the product of this operation by F'0 (Mosaic Analysis Software). (MOV 7949 kb)
Temporal down sampling of video 6.
If the frame rate used to collect the raw data files is greater than what is needed to detect distinct Ca2+ transients from individual cells, down sample the movie in the temporal domain to reduce the file size and speed up the rate of data analysis. In this example, the data was acquired at 15 Hz and binned by a factor of 3, converting the sampling rate to 5 Hz (Mosaic Analysis Software). (MOV 6868 kb)
Visualization of in vivo Ca2+ signals from the cortex of a freely behaving mouse.
A 1 mm diameter GRIN lens was implanted above the barrel field region of the somatosensory cortex of a wild type male mouse (AAVdj-CamkII-GCaMP6f, titer: 5.6 × 1012, 1:8 dilution, UNC vector core). in vivo Ca2+ transients (left) were visualized through a GRIN lens with a miniature, fluorescence microscope secured to the head of a freely behaving mouse (right). Notice that the location of detected transients remains stable even while the mouse grooms his facial region near the point of microscope attachment. Ca2+ imaging movie was processed with Mosaic Analysis Software (Spatial bin by 4, motion corrected, cropped, conversion to ΔF/F format, temporal bin by 3). All procedures were approved by UNC IACUC. (MOV 9075 kb)
Visualization of subcortical Ca2+ signals in a freely behaving mouse.
A 1 mm diameter GRIN lens was implanted above the dentate gyrus of a wild type male mouse (AAVdj-CamkII-GCaMP6m, titer: 7.0 × 1012, 1:1 dilution, Stanford). in vivo Ca2+ transients were visualized through the implanted GRIN lens with a miniature, fluorescence microscope secured to the head of the mouse. The Ca2+ imaging movie was processed with Mosaic Analysis Software (Spatial bin by 4, motion corrected, cropped, conversion to ΔF/F format, temporal bin by 3). All procedures were approved by UNC IACUC. (MOV 8822 kb)
Visualization of deep brain Ca2+ signals in a freely behaving mouse.
A 0.5 mm diameter GRIN lens was implanted above the lateral hypothalamus (LH) of a VGAT-Cre male mouse following microinjection of a genetically encoded Ca2+ indicator (AAVdj-EF1a-DIO-DIO-GCaMP6m, titer 9.7 × 1012, 1:2 dilution, Stanford) into the LH. Ca2+ transients within GABAergic LH neurons were visualized through the implanted GRIN lens with a miniature, fluorescence microscope secured to the head of the mouse. Imaging data was acquired at 15 Hz. Data shown is from an unprocessed movie. All procedures were approved by UNC IACUC. (MOV 26010 kb)
Supplementary Data 1
Zip file that contains analysis GUI for the analysis of Ca2+ imaging data in relation to temporally defined events as well as a function to plot the activity of individual cells in relation to their location in space. (ZIP 44 kb)
Rights and permissions
About this article
Cite this article
Resendez, S., Jennings, J., Ung, R. et al. Visualization of cortical, subcortical and deep brain neural circuit dynamics during naturalistic mammalian behavior with head-mounted microscopes and chronically implanted lenses. Nat Protoc 11, 566–597 (2016). https://doi.org/10.1038/nprot.2016.021
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.021
This article is cited by
-
Top-down control of flight by a non-canonical cortico-amygdala pathway
Nature (2024)
-
The Cousa objective: a long-working distance air objective for multiphoton imaging in vivo
Nature Methods (2024)
-
Activity-dependent organization of prefrontal hub-networks for associative learning and signal transformation
Nature Communications (2023)
-
Construction and use of an adaptive optics two-photon microscope with direct wavefront sensing
Nature Protocols (2023)
-
Unraveling the mechanisms of deep-brain stimulation of the internal capsule in a mouse model
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.