Abstract
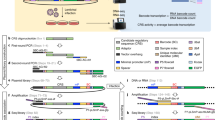
This protocol provides a rapid, streamlined and scalable strategy to systematically scan genomic regions for the presence of transcriptional regulatory regions that are active in a specific cell type. It creates genomic tiles spanning a region of interest that are subsequently cloned by recombination into a luciferase reporter vector containing the simian virus 40 promoter. Tiling clones are transfected into specific cell types to test for the presence of transcriptional regulatory regions. The protocol includes testing of different single-nucleotide polymorphism (SNP) alleles to determine their effect on regulatory activity. This procedure provides a systematic framework for identifying candidate functional SNPs within a locus during functional analysis of genome-wide association studies. This protocol adapts and combines previous well-established molecular biology methods to provide a streamlined strategy, based on automated primer design and recombinational cloning, allowing one to rapidly go from a genomic locus to a set of candidate functional SNPs in 8 weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Manolio, T.A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 363, 166–176 (2010).
Lewis, C.M. & Knight, J. Introduction to genetic association studies. Cold Spring Harb. Protoc. 2012, 297–306 (2012).
Freedman, M.L. et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat. Genet. 43, 513–518 (2011).
Edwards, S.L., Beesley, J., French, J.D. & Dunning, A.M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93, 779–797 (2013).
Monteiro, A.N. & Freedman, M.L. Lessons from postgenome-wide association studies: functional analysis of cancer predisposition loci. J. Intern. Med. 274, 414–424 (2013).
Tang, W. et al. Mapping of the UGT1A locus identifies an uncommon coding variant that affects mRNA expression and protects from bladder cancer. Hum. Mol. Genet. 21, 1918–1930 (2012).
Maurano, M.T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Sakoda, L.C., Jorgenson, E. & Witte, J.S. Turning of COGS moves forward findings for hormonally mediated cancers. Nat. Genet. 45, 345–348 (2013).
Chung, C.C., Magalhaes, W.C., Gonzalez-Bosquet, J. & Chanock, S.J. Genome-wide association studies in cancer—current and future directions. Carcinogenesis 31, 111–120 (2010).
Carey, M. & Smale, S.T. in Transcriptional Regulation in Eukaryotes: Concepts, Strategies and Techniques (Cold Spring Harbor Laboratory Press, 1999).
Pharoah, P.D. et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 45, 362–370 (2013).
Baskin, R. et al. Functional analysis of the 11q23.3 glioma susceptibility locus implicates PHLDB1 and DDX6 in glioma susceptibility. Sci. Rep. 5, 17367 (2015).
Neph, S. et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90 (2012).
Thurman, R.E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Sanyal, A., Lajoie, B.R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012).
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012).
Gerstein, M.B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012).
Andersson, R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014).
FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
Stergachis, A.B. et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science 342, 1367–1372 (2013).
Mansour, M.R. et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346, 1373–1377 (2014).
Melnikov, A. et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30, 271–277 (2012).
Arnold, C.D. et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077 (2013).
Plank, J.L. & Dean, A. Enhancer function: mechanistic and genome-wide insights come together. Mol. Cell 55, 5–14 (2014).
van Arensbergen, J., van Steensel, B. & Bussemaker, H.J. In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 24, 695–702 (2014).
Carlson, C.S. et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 74, 106–120 (2004).
Hazelett, D.J. et al. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 10, e1004102 (2014).
French, J.D. et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet. 92, 489–503 (2013).
Boyle, A.P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Coetzee, S.G., Rhie, S.K., Berman, B.P., Coetzee, G.A. & Noushmehr, H. FunciSNP: an R/Bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 40, e139 (2012).
Blackwood, E.M. & Kadonaga, J.T. Going the distance: a current view of enhancer action. Science 281, 60–63 (1998).
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Khoury, G. & Gruss, P. Enhancer elements. Cell 33, 313–314 (1983).
Braman, J., Papworth, C. & Greener, A. Site-directed mutagenesis using double-stranded plasmid DNA templates. Methods Mol. Biol. 57, 31–44 (1996).
Machiela, M.J. & Chanock, S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015).
Millot, G.A. et al. A guide for functional analysis of BRCA1 variants of uncertain significance. Hum. Mutat. 33, 1526–1537 (2012).
Ikram, M.K. et al. Four novel loci (19q13, 6q24, 12q24 and 5q14) influence the microcirculation in vivo. PLoS Genet. 6, e1001184 (2010).
Acknowledgements
This work was supported by the US National Institutes of Health (NIH) Genetic Association and Mechanisms in Oncology (GAME-ON) through the National Cancer Institute (NCI) U19 award (no. CA148112), the Ovarian Cancer Research Foundation (no. 258807), the Phi Beta Psi Foundation and in part by the Molecular Genomics Core Facilities at the Moffitt Cancer Center through its NCI Cancer Center Support Grant (grant no. P30-CA76292). M.B. is an ARCS (Achievement Rewards for College Scientists) fellow and a recipient of the Ruth L. Kirschstein National Research Service Award (no. F31 CA165528). R.B. is a trainee on an NIH R25T award (no. CA147832). R.S.C. is supported by a fellowship from the Brazilian National Council for Scientific and Technological Development (CNPq). We thank A. Valle, P. Cilas Jr., B. Reid and X. Li for technical assistance.
Author information
Authors and Affiliations
Contributions
M.B., A.G., R.S.C., N.T.W. and A.N.A.M. conceived the project and designed the experiments. M.B., G.M.-F., A.G., R.B. and R.S.C. performed the experiments. M.B., G.M.-F., A.G., R.B., R.S.C., M.A.C., N.T.W. and A.N.A.M. performed the analysis and contributed to the discussion and overall data interpretation. M.B., G.M.-F., A.G. and A.N.A.M. wrote the paper. All authors provided intellectual input and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Steps to generate a BED file containing SNP coordinates from LDlink or SNAP Proxy. Contains step-by-step examples of data generated from LDlink or SNA Proxy and formatted for uploading onto the UCSC Human Genome Browser. (XLSX 718 kb)
Rights and permissions
About this article
Cite this article
Buckley, M., Gjyshi, A., Mendoza-Fandiño, G. et al. Enhancer scanning to locate regulatory regions in genomic loci. Nat Protoc 11, 46–60 (2016). https://doi.org/10.1038/nprot.2015.136
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.136
This article is cited by
-
Assessment of small in-frame indels and C-terminal nonsense variants of BRCA1 using a validated functional assay
Scientific Reports (2022)
-
A 16q22.1 variant confers susceptibility to colorectal cancer as a distal regulator of ZFP90
Oncogene (2020)
-
Functional Landscape of Common Variants Associated with Susceptibility to Epithelial Ovarian Cancer
Current Epidemiology Reports (2020)
-
A transcriptome-wide association study of high-grade serous epithelial ovarian cancer identifies new susceptibility genes and splice variants
Nature Genetics (2019)
-
Two novel cyclic hexapeptides from the genetically engineered Actinosynnema pretiosum
Applied Microbiology and Biotechnology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.