Abstract
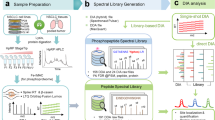
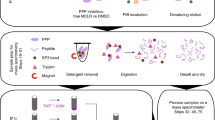
Mass spectrometry–based phosphoproteomic analysis is a powerful method for gaining a global, unbiased understanding of cellular signaling. Its accuracy and comprehensiveness stands or falls with the quality and choice of the applied phosphopeptide prefractionation strategy. This protocol covers a powerful but simple and rapid strategy for phosphopeptide prefractionation. The combinatorial use of two distinct chromatographic techniques that address the inverse physicochemical properties of peptides allows for superior fractionation efficiency of multiple phosphorylated peptides. In the first step, multiphosphorylated peptides are separated according to the number of negatively charged phosphosites by electrostatic repulsion-hydrophilic interaction chromatography (ERLIC). A subsequent strong cation exchange (SCX) step separates mostly singly phosphorylated peptides in the ERLIC flow-through according to their positive charge. The presented strategy is inexpensive and adaptable to large and small amounts of starting material, and it allows highly multiplexed sample preparation. Because of its implementation as solid-phase extraction, the entire workflow takes only 2 h to complete.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sharma, K. et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 (2014).
Rigbolt, K.T. & Blagoev, B. Quantitative phosphoproteomics to characterize signaling networks. Semin. Cell Dev. Biol. 23, 863–871 (2012).
Rigbolt, K.T. et al. Characterization of early autophagy signaling by quantitative phosphoproteomics. Autophagy 10, 356–371 (2014).
Olsen, J.V. et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 (2010).
Kuroda, I., Shintani, Y., Motokawa, M., Abe, S. & Furuno, M. Phosphopeptide-selective column-switching RP-HPLC with a titania precolumn. Anal. Sci. 20, 1313–1319 (2004).
Thingholm, T.E., Jorgensen, T.J., Jensen, O.N. & Larsen, M.R. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protoc. 1, 1929–1935 (2006).
Di Palma, S., Hennrich, M.L., Heck, A.J. & Mohammed, S. Recent advances in peptide separation by multidimensional liquid chromatography for proteome analysis. J. Proteomics 75, 3791–3813 (2012).
Beausoleil, S.A. et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 101, 12130–12135 (2004).
Pinkse, M.W., Uitto, P.M., Hilhorst, M.J., Ooms, B. & Heck, A.J. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 (2004).
Zarei, M., Sprenger, A., Gretzmeier, C. & Dengjel, J. Combinatorial use of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) and strong cation exchange (SCX) chromatography for in-depth phosphoproteome analysis. J. Proteome Res. 11, 4269–4276 (2012).
Alpert, A.J. Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal. Chem. 80, 62–76 (2008).
Loroch, S., Schommartz, T., Brune, W., Zahedi, R.P. & Sickmann, A. Multidimensional electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) for quantitative analysis of the proteome and phosphoproteome in clinical and biomedical research. Biochim. Biophys. Acta 1854, 460–468 (2015).
Loroch, S., Zahedi, R.P. & Sickmann, A. Highly sensitive phosphoproteomics by tailoring solid-phase extraction to electrostatic repulsion-hydrophilic interaction chromatography. Anal. Chem. 87, 1596–1604 (2015).
Hao, P. et al. Enhanced separation and characterization of deamidated peptides with RP-ERLIC-based multidimensional chromatography coupled with tandem mass spectrometry. J. Proteome Res. 11, 1804–1811 (2012).
Alpert, A.J., Hudecz, O. & Mechtler, K. Anion-exchange chromatography of phosphopeptides: weak anion exchange versus strong anion exchange and anion-exchange chromatography versus electrostatic repulsion-hydrophilic interaction chromatography. Anal. Chem. 87, 4704–4711 (2015).
Stuart, S.A. et al. A phosphoproteomic comparison of B-RAFV600E and MKK1/2 inhibitors in melanoma cells. Mol. Cell. Proteomics 14, 1599–1615 (2015).
Zarei, M., Sprenger, A., Metzger, F., Gretzmeier, C. & Dengjel, J. Comparison of ERLIC-TiO2, HILIC-TiO2, and SCX-TiO2 for global phosphoproteomics approaches. J. Proteome Res. 10, 3474–3483 (2011).
Zarei, M., Sprenger, A., Gretzmeier, C. & Dengjel, J. Rapid combinatorial ERLIC-SCX solid-phase extraction for in-depth phosphoproteome analysis. J. Proteome Res. 12, 5989–5995 (2013).
Rush, J. et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 (2005).
Blagoev, B. et al. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21, 315–318 (2003).
Edbauer, D. et al. Identification and characterization of neuronal mitogen-activated protein kinase substrates using a specific phosphomotif antibody. Mol. Cell. Proteomics 8, 681–695 (2009).
Matheron, L., van den Toorn, H., Heck, A.J. & Mohammed, S. Characterization of biases in phosphopeptide enrichment by Ti4+-immobilized metal affinity chromatography and TiO2 using a massive synthetic library and human cell digests. Anal. Chem. 86, 8312–8320 (2014).
McNulty, D.E. & Annan, R.S. Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol. Cell. Proteomics 7, 971–980 (2008).
Batth, T.S., Francavilla, C. & Olsen, J.V. Off-line high-pH reversed-phase fractionation for in-depth phosphoproteomics. J. Proteome Res. 13, 6176–6186 (2014).
Andersson, L. & Porath, J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 154, 250–254 (1986).
Beltran, L. & Cutillas, P.R. Advances in phosphopeptide enrichment techniques for phosphoproteomics. Amino Acids 43, 1009–1024 (2012).
Larsen, M.R., Thingholm, T.E., Jensen, O.N., Roepstorff, P. & Jorgensen, T.J. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 (2005).
Tsai, C.F. et al. Sequential phosphoproteomic enrichment through complementary metal-directed immobilized metal ion affinity chromatography. Anal. Chem. 86, 685–693 (2014).
Bodenmiller, B., Mueller, L.N., Mueller, M., Domon, B. & Aebersold, R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat. Methods 4, 231–237 (2007).
Kettenbach, A.N. & Gerber, S.A. Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal. Chem. 83, 7635–7644 (2011).
Montoya, A., Beltran, L., Casado, P., Rodriguez-Prados, J.C. & Cutillas, P.R. Characterization of a TiO2 enrichment method for label-free quantitative phosphoproteomics. Methods 54, 370–378 (2011).
Li, Q.R., Ning, Z.B., Tang, J.S., Nie, S. & Zeng, R. Effect of peptide-to-TiO2 beads ratio on phosphopeptide enrichment selectivity. J. Proteome Res. 8, 5375–5381 (2009).
Wisniewski, J.R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Lin, Y. et al. Sodium-deoxycholate-assisted tryptic digestion and identification of proteolytically resistant proteins. Anal. Biochem. 377, 259–266 (2008).
Rogers, L.D., Fang, Y. & Foster, L.J. An integrated global strategy for cell lysis, fractionation, enrichment and mass spectrometric analysis of phosphorylated peptides. Mol. Biosyst. 6, 822–829 (2010).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Cox, J. & Mann, M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13 (suppl. 16) S12 (2012).
Rigbolt, K.T., Vanselow, J.T. & Blagoev, B. GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol. Cell. Proteomics 10, O110.007450 (2011).
Zhou, H. et al. Enhancing the identification of phosphopeptides from putative basophilic kinase substrates using Ti (IV) based IMAC enrichment. Mol. Cell. Proteomics 10, M110.006452 (2011).
Colaert, N., Helsens, K., Martens, L., Vandekerckhove, J. & Gevaert, K. Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 (2009).
Acknowledgements
The research leading to these results has received funding from the Excellence Initiative of the German Federal and State Governments through FRIAS and BIOSS, from the German Research Foundation (DFG) via SFB1140, KIDGEM Project C02, and project DE 1757/2-1. We thank Eksigent/AB Sciex for the generous gift of a 2D nanoLC and R. van Soest for his technical advice on maintenance and troubleshooting.
Author information
Authors and Affiliations
Contributions
M.Z. and A.S. set up the protocol and performed the experiments. M.R. performed data analysis. J.D. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Zarei, M., Sprenger, A., Rackiewicz, M. et al. Fast and easy phosphopeptide fractionation by combinatorial ERLIC-SCX solid-phase extraction for in-depth phosphoproteome analysis. Nat Protoc 11, 37–45 (2016). https://doi.org/10.1038/nprot.2015.134
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.134
This article is cited by
-
Global kinome profiling reveals DYRK1A as critical activator of the human mitochondrial import machinery
Nature Communications (2021)
-
Magnetite nanoparticles coated with chitosan and polyethylenimine as anion exchanger for sorptive enrichment of phosphopeptides
Microchimica Acta (2019)
-
Phytic acid functionalized Fe3O4 nanoparticles loaded with Ti(IV) ions for phosphopeptide enrichment in mass spectrometric analysis
Microchimica Acta (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.