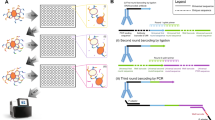
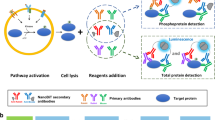
Abstract
This protocol describes a highly reproducible antibody-based method that provides protein level and phosphorylation status information from nanogram quantities of protein cell lysate. Nanocapillary isoelectric focusing (cIEF) combines with UV-activated linking chemistry to detect changes in phosphorylation status. As an example application, we describe how to detect changes in response to tyrosine kinase inhibitors (TKIs) in the phosphorylation status of the adaptor protein CrkL, a major substrate of the oncogenic tyrosine kinase BCR-ABL in chronic myeloid leukemia (CML), using highly enriched CML stem cells and mature cell populations in vitro. This protocol provides a 2.5 pg/nl limit of protein detection (<0.2% of a stem cell sample containing <104 cells). Additional assays are described for phosphorylated tyrosine 207 (pTyr207)-CrkL and the protein tyrosine phosphatase PTPRC/CD45; these assays were developed using this protocol and applied to CML patient samples. This method is of high throughput, and it can act as a screen for in vitro cancer stem cell response to drugs and novel agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
O'Neill, R.A. et al. Isoelectric focusing technology quantifies protein signaling in 25 cells. Proc. Natl. Acad. Sci. USA 103, 16153–16158 (2006).
Fan, A.C. et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat. Med. 15, 566–571 (2009).
Maiso, P. et al. Defining the role of TORC1/2 in multiple myeloma. Blood 118, 6860–6870 (2011).
Kentsis, A. et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat. Med. 18, 1118–1122 (2012).
Williamson, A.J.K. et al. A specific PTPRC/CD45 phosphorylation event governed by stem cell chemokine CXCL12 regulates primitive hematopoietic cell motility. Mol. Cell. Proteom. 12, 3319–3329 (2013).
Brown, S. et al. Monocyte-derived dendritic cells from chronic myeloid leukaemia have abnormal maturation and cytoskeletal function that is associated with defective localisation and signalling by normal ABL1 protein. Eur. J. Haematol. 93, 96–102 (2014).
Al-Hajj, M., Wicha, M.S., Benito-Hernandez, A., Morrison, S.J. & Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 100, 3983–3988 (2003).
Li, C. et al. Identification of pancreatic cancer stem cells. Cancer Res. 67, 1030–1037 (2007).
Prince, M. et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 104, 973–981 (2007).
Mitelman, F., Johansson, B. & Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7, 233–245 (2007).
Sanz, M., Burnett, A., Lo-Coco, F. & Lowenberg, B. FLT3 inhibition as a targeted therapy for acute myeloid leukemia. Curr. Opin. Oncol. 21, 594–600 (2009).
Verstovsek, S. Therapeutic potential of JAK2 inhibitors. Hematol. Am. Soc. Hematol. Educ. Prog. 2009, 636–642 (2009).
Rowley, J.D. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243, 290–293 (1973).
Groffen, J. et al. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell 36, 93–99 (1984).
Druker, B.J. et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl–positive cells. Nat. Med. 2, 561–566 (1996).
Gambacorti-Passerini, C. et al. Inhibition of the ABL kinase activity blocks the proliferation of BCR/ABL+ leukemic cells and induces apoptosis. Blood Cells Mol. Dis. 23, 380–394 (1997).
Hamilton, A. et al. BCR-ABL activity and its response to drugs can be determined in CD34+ CML stem cells by CrkL phosphorylation status using flow cytometry. Leukemia 20, 1035–1039 (2006).
Copland, M. et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood 107, 4532–4539 (2006).
Hochhaus, A. et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia 23, 1054–1061 (2009).
Hochhaus, A. et al. Favorable long-term follow-up results over 6 years for response, survival, and safety with imatinib mesylate therapy in chronic-phase chronic myeloid leukemia after failure of interferon-α treatment. Blood 111, 1039–1043 (2008).
O'Hare, T., Zabriskie, M.S., Eiring, A.M. & Deininger, M.W. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat. Rev. Cancer 12, 513–526 (2012).
Graham, S.M. et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571. Blood 99, 319–325 (2002).
Holtz, M.S. Imatinib mesylate (STI571) inhibits growth of primitive malignant progenitors in chronic myelogenous leukemia through reversal of abnormally increased proliferation. Blood 99, 3792–3800 (2002).
Mahon, F.X. et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 11, 1029–1035 (2010).
Chu, S. et al. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood 118, 5565–5572 (2011).
Rousselot, P. et al. Imatinib mesylate discontinuation in patients with chronic myelogenous leukemia in complete molecular remission for more than 2 years. Blood 109, 58–60 (2007).
Druker, B.J. et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 355, 2408–2417 (2006).
Addona, T.A. et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 (2009).
Hjelle, S.M. et al. Clinical proteomics of myeloid leukemia. Genome Med. 2, 41 (2010).
de Jong, R., ten Hoeve, J., Heisterkamp, N. & Groffen, J. Tyrosine 207 in CRKL is the BCR/ABL phosphorylation site. Oncogene 14, 507–513 (1997).
Seo, J.H. et al. A specific need for CRKL in p210BCR-ABL-induced transformation of mouse hematopoietic progenitors. Cancer Res. 70, 7325–7335 (2010).
Konig, H. et al. Effects of dasatinib on SRC kinase activity and downstream intracellular signaling in primitive chronic myelogenous leukemia hematopoietic cells. Cancer Res. 68, 9624–9633 (2008).
Oda, T. et al. Crkl is the major tyrosine-phosphorylated protein in neutrophils from patients with chronic myelogenous leukemia. J. Biol. Chem. 269, 22925–22928 (1994).
Liu, X. et al. Constrained selected reaction monitoring: quantification of selected post-translational modifications and protein isoforms. Methods 61, 304–312 (2013).
Maxwell, S.A. et al. Analysis of p210bcr-abl tyrosine protein kinase activity in various subtypes of Philadelphia chromosome-positive cells from chronic myelogenous leukemia patients. Cancer Res. 47, 1731–1739 (1987).
Copland, M., Hamilton, A. & Holyoake, T.L. Response: conventional western blotting techniques will not reliably quantify p210 BCR-ABL. Blood 109, 1336 (2007).
Bendall, S.C. & Nolan, G.P. From single cells to deep phenotypes in cancer. Nat. Biotechnol. 30, 639–647 (2012).
Irish, J.M. et al. Single-cell profiling of potentiated phospho-protein networks in cancer cells. Cell 118, 217–228 (2004).
Bendall, S.C., Nolan, G.P., Roederer, M. & Chattopadhyay, P.K. A deep profiler's guide to cytometry. Trends Immunol. 33, 323–332 (2012).
Hamilton, A., Alhashimi, F., Myssina, S., Jorgensen, H.G. & Holyoake, T.L. Optimization of methods for the detection of BCR-ABL activity in Philadelphia-positive cells. Exp. Hematol. 37, 395–401 (2009).
Protein Simple, Inc. Application Brief, Vol. 1023 http://www.proteinsimple.com/technical_library.html?product=simplewestern&def_list=list (Protein Simple, 2010).
Hamilton, A. et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood 119, 1501–1511 (2012).
Hornbeck, P.V. et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 (2012).
Konopka, J.B., Watanabe, S.M. & Witte, O.N. An alteration of the human c-abl protein in K562 leukemia cells unmasks associated tyrosine kinase activity. Cell 37, 1035–1042 (1984).
Golub, T.R., Barker, G.F., Lovett, M. & Gilliland, D.G. Fusion of PDGF receptor β to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 77, 307–316 (1994).
Cools, J. et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N. Engl. J. Med. 348, 1201–1214 (2003).
Morris, S.W. et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science 263, 1281–1284 (1994).
Nagata, H. et al. Identification of a point mutation in the catalytic domain of the proto-oncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc. Natl. Acad. Sci. USA 92, 10560–10564 (1995).
Ziegler, S. et al. A novel protein kinase D phosphorylation site in the tumor suppressor Rab interactor 1 is critical for coordination of cell migration. Mol. Biol. Cell 22, 570–580 (2011).
Shah, N.P. et al. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell 14, 485–493 (2008).
Young, M.A. et al. Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680. Cancer Res. 66, 1007–1014 (2006).
Zipfel, P.A., Zhang, W., Quiroz, M. & Pendergast, A.M. Requirement for Abl kinases in T cell receptor signaling. Curr. Biol. 14, 1222–1231 (2004).
Uemura, N. & Griffin, J.D. The adapter protein Crkl links Cbl to C3G after integrin ligation and enhances cell migration. J. Biol. Chem. 274, 37525–37532 (1999).
Senechal, K., Heaney, C., Druker, B. & Sawyers, C.L. Structural requirements for function of the Crkl adapter protein in fibroblasts and hematopoietic cells. Mol. Cell Biol. 18, 5082–5090 (1998).
Nichols, G.L. et al. Identification of CRKL as the constitutively phosphorylated 39-kDa tyrosine phosphoprotein in chronic myelogenous leukemia cells. Blood 84, 2912–2918 (1994).
ten Hoeve, J., Arlinghaus, R.B., Guo, J.Q., Heisterkamp, N. & Groffen, J. Tyrosine phosphorylation of CRKL in Philadelphia+ leukemia. Blood 84, 1731–1736 (1994).
Acknowledgements
This work was supported by Leukaemia and Lymphoma Research UK (08004 to F.P., M.T.S. and T.L.H.), Cancer Research UK (C11074/A11008 to F.P., M.T.S. and T.L.H.) and the Chief Scientist Office (CZB/4/690 to F.P., M.T.S. and T.L.H.), and also by the Glasgow and Manchester Experimental Cancer Medicine Centres, which are funded by Cancer Research UK and the Chief Scientist Office in Scotland.
Author information
Authors and Affiliations
Contributions
A.D.W. and T.L.H. devised the study; M.A.-O. performed the experimental and study design and wrote the manuscript with A.D.W.; A.P. contributed to study design and instrument setup; F.P. and M.T.S. prepared clinical material; A.J.W. provided the Src inhibitor; and all authors contributed experimental methods and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 CD45 assay development.
Representative trace for total-CD45 (black) and pCD45 (red). b, LOD curve for CD45. c, UV titration for CD45 using total antibody, error bars S.E.M (n=3 technical replicate).
Supplementary Figure 2 Lambda phosphatase treatment of HL60 and K562 cell lysates.
Assay plate design and results created using Compass software assay and analysis perspectives respectively. a, layout of samples, antibodies and reagents in 384-well plate format b, detailed description of samples and antibodies applied to plate designed in (a). c, representative differences in total CrkL spectra resulting from lambda phosphatase (λp) treatment, K562-NDC (black), K562-NDC-λp (green), K562-IM 5µM-λp (blue) and K562-DAS 150nM-λp (red).
Supplementary Figure 3 Spectra mismatching and poor ladder detection.
Assay data showing total CrkL antibody probed against K562 cell lysate with no TKI reatment. Spectra shown represent technical replicates from the same sample. a, mismatched data example; correct standard ladder pI associations applied to the black trace, and incorrect associations applied to the blue trace. Red dotted lines indicate matching peaks and the extent of peak offset in the blue trace as a result of poor ladder detection. b, manual alteration of this error results in perfect alignment.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 927 kb)
Poor separation; sample salt content > 150 mM.
Video showing the effect on protein separation with cIEF when using clinical samples prepared in a high ionic strength buffer (n = 4 biological samples, loaded in triplicate). (MOV 1916 kb)
Good separation; sample salt content <150 mM.
Video showing the effect on protein separation with cIEF when using clinical samples prepared in a low ionic strength buffer (n = 4 biological samples, loaded in triplicate). (MOV 1402 kb)
Rights and permissions
About this article
Cite this article
Aspinall-O'Dea, M., Pierce, A., Pellicano, F. et al. Antibody-based detection of protein phosphorylation status to track the efficacy of novel therapies using nanogram protein quantities from stem cells and cell lines. Nat Protoc 10, 149–168 (2015). https://doi.org/10.1038/nprot.2015.007
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.007
This article is cited by
-
Detection of the Cell Cycle-Regulated Negative Feedback Phosphorylation of Mitogen-Activated Protein Kinases in Breast Carcinoma using Nanofluidic Proteomics
Scientific Reports (2018)
-
Proteomic analysis of JAK2V617F-induced changes identifies potential new combinatorial therapeutic approaches
Leukemia (2017)
-
Decreased Glycogen Content Might Contribute to Chronic Stress-Induced Atrophy of Hippocampal Astrocyte volume and Depression-like Behavior in Rats
Scientific Reports (2017)
-
Human keratinocytes have two interconvertible modes of proliferation
Nature Cell Biology (2016)
-
Capillary nano-immunoassays: advancing quantitative proteomics analysis, biomarker assessment, and molecular diagnostics
Journal of Translational Medicine (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.