Abstract
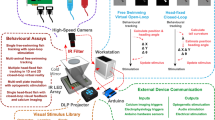
This article describes a method to quantify the movements of larval zebrafish in multiwell plates, using the open-source MATLAB applications LSRtrack and LSRanalyze. The protocol comprises four stages: generation of high-quality, flatly illuminated video recordings with exposure settings that facilitate object recognition; analysis of the resulting recordings using tools provided in LSRtrack to optimize tracking accuracy and motion detection; analysis of tracking data using LSRanalyze or custom MATLAB scripts; and implementation of validation controls. The method is reliable, automated and flexible, requires <1 h of hands-on work for completion once optimized and shows excellent signal:noise characteristics. The resulting data can be analyzed to determine the following: positional preference; displacement, velocity and acceleration; and duration and frequency of movement events and rest periods. This approach is widely applicable to the analysis of spontaneous or stimulus-evoked zebrafish larval neurobehavioral phenotypes resulting from a broad array of genetic and environmental manipulations, in a multiwell plate format suitable for high-throughput applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Mahmood, F., Fu, S. & Cooke, J. et al. A zebrafish model of CLN2 disease is deficient in tripeptidyl peptidase 1 and displays progressive neurodegeneration accompanied by a reduction in proliferation. Brain 136 (Part 5): 1488–1507 (2013).
Milanese, C., Sager, J.J. & Bai, Q. et al. Hypokinesia and reduced dopamine levels in zebrafish lacking β- and γ1-synucleins. J. Biol. Chem. 287, 2971–2983 (2012).
Elbaz, I., Yelin-Bekerman, L. & Nicenboim, J. et al. Genetic ablation of hypocretin neurons alters behavioral state transitions in zebrafish. J. Neurosci. 32, 12961–12972 (2012).
Rihel, J., Prober, D.A. & Arvanites, A. et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 327, 348–351 (2010).
Sallinen, V., Torkko, V. & Sundvik, M. et al. MPTP and MPP+ target specific aminergic cell populations in larval zebrafish. J. Neurochem. 108, 719–731 (2009).
Kokel, D., Bryan, J. & Laggner, C. et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat. Chem. Biol. 6, 231–237 (2010).
Zon, L.I. & Peterson, R.T. In vivo drug discovery in the zebrafish. Nat. Rev. Drug. Discov. 4, 35–44 (2005).
Farrell, T.C., Cario, C.L. & Milanese, C. et al. Evaluation of spontaneous propulsive movement as a screening tool to detect rescue of Parkinsonism phenotypes in zebrafish models. Neurobiol. Dis. 44, 9–18 (2011).
Prober, D.A., Rihel, J. & Onah, A.A. et al. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J. Neurosci. 26, 13400–13410 (2006).
Burgess, H.A. & Granato, M. Modulation of locomotor activity in larval zebrafish during light adaptation. J. Exp. Biol. 210 (Part 14): 2526–2539 (2007).
Cario, C.L., Farrell, T.C. & Milanese, C. et al. Automated measurement of zebrafish larval movement. J. Physiol. 589 (Part 15): 3703–3708 (2011).
Sager, J.J., Torres, G.E. & Burton, E.A. The zebrafish homologue of the human DYT1 dystonia gene is widely expressed in CNS neurons but non-essential for early motor system development. PLoS ONE 7, e45175 (2012).
Richendrfer, H., Pelkowski, S.D. & Colwill, R.M. et al. On the edge: pharmacological evidence for anxiety-related behavior in zebrafish larvae. Behav. Brain Res. 228, 99–106 (2012).
Fontaine, E., Lentink, D. & Kranenbarg, S. et al. Automated visual tracking for studying the ontogeny of zebrafish swimming. J. Exp. Biol. 211 (Part 8): 1305–1316 (2008).
Creton, R. Automated analysis of behavior in zebrafish larvae. Behav. Brain Res. 203, 127–136 (2009).
Martineau, P.R. & Mourrain, P. Tracking zebrafish larvae in group—status and perspectives. Methods 62, 292–303 (2013).
Burgess, H.A. & Granato, M. Sensorimotor gating in larval zebrafish. J. Neurosci. 27, 4984–4994 (2007).
Acknowledgements
Development of LSRtrack and the current protocol was supported by research grants from the National Institute of Neurological Disorders and Stroke (NINDS) (NS080881 and NS058369), the National Institute of Environmental Health Sciences (NIEHS) (ES022644), the Pittsburgh Foundation (M2005-0071) and the Society for Progressive Supranuclear Palsy (468-08). Y.Z. is a Tsinghua Scholar at the University of Pittsburgh.
Author information
Authors and Affiliations
Contributions
Y.Z., C.L.C. and E.A.B. designed and wrote the software; Y.Z., R.T.C., Q.B. and E.A.B. developed the experimental protocol; Y.Z., R.T.C. and Q.B. carried out the experiments; and Y.Z., R.T.C. and E.A.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Methods
Instructions and drawings are shown for construction of optional equipment used in the protocol. (PDF 508 kb)
Supplementary Software
Software is adapted from Cario, C.L., Farrell, T.C., Milanese, C. & Burton, E.A. Automated measurement of zebrafish larval movement. J. Physiol. 589 3703–3708 (2011) with permission from Wiley (© 2011 The Authors. Journal compilation © 2011 The Physiological Society). (ZIP 46 kb)
Rights and permissions
About this article
Cite this article
Zhou, Y., Cattley, R., Cario, C. et al. Quantification of larval zebrafish motor function in multiwell plates using open-source MATLAB applications. Nat Protoc 9, 1533–1548 (2014). https://doi.org/10.1038/nprot.2014.094
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.094
This article is cited by
-
The medaka mutant deficient in eyes shut homolog exhibits opsin transport defects and enhanced autophagy in retinal photoreceptors
Cell and Tissue Research (2023)
-
Zebrafish tracking using YOLOv2 and Kalman filter
Scientific Reports (2021)
-
Embryonic exposure to ethanol increases the susceptibility of larval zebrafish to chemically induced seizures
Scientific Reports (2018)
-
Action sequencing in the spontaneous swimming behavior of zebrafish larvae - implications for drug development
Scientific Reports (2017)
-
Automatic multiple zebrafish larvae tracking in unconstrained microscopic video conditions
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.