Abstract
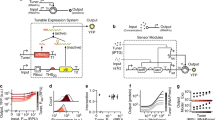
Memory and logic are central to complex state-dependent computing, and state-dependent behaviors are a feature of natural biological systems. Recently, we created a platform for integrated logic and memory by using synthetic gene circuits, and we demonstrated the implementation of all two-input logic gates with memory in living cells. Here we provide a detailed protocol for the construction of two-input Boolean logic functions with concomitant DNA-based memory. This technology platform allows for straightforward assembly of integrated logic-and-memory circuits that implement desired behaviors within a couple of weeks. It should enable the encoding of advanced computational operations in living cells, including sequential-logic and biological-state machines, for a broad range of applications in biotechnology, basic science and biosensing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Cheng, A.A. & Lu, T.K. Synthetic biology: an emerging engineering discipline. Annu. Rev. Biomed. Eng. 14, 155–178 (2012).
Lu, T.K., Khalil, A.S. & Collins, J.J. Next-generation synthetic gene networks. Nat. Biotechnol. 27, 1139–1150 (2009).
Tamsir, A., Tabor, J.J. & Voigt, C.A. Robust multicellular computing using genetically encoded NOR gates and chemical 'wires'. Nature 469, 212–215 (2011).
Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 13, 455–468 (2012).
Auslander, S., Auslander, D., Muller, M., Wieland, M. & Fussenegger, M. Programmable single-cell mammalian biocomputers. Nature 487, 123–127 (2012).
Xie, Z., Wroblewska, L., Prochazka, L., Weiss, R. & Benenson, Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333, 1307–1311 (2011).
Gardner, T.S., Cantor, C.R. & Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Becskei, A., Seraphin, B. & Serrano, L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 20, 2528–2535 (2001).
Ajo-Franklin, C.M. et al. Rational design of memory in eukaryotic cells. Gene Dev. 21, 2271–2276 (2007).
Nevozhay, D., Adams, R.M., Van Itallie, E., Bennett, M.R. & Balazsi, G. Mapping the environmental fitness landscape of a synthetic gene circuit. PLoS Comput. Biol. 8, e1002480 (2012).
Siuti, P., Yazbek, J. & Lu, T.K. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 31, 448–452 (2013).
Wang, Y., Yau, Y.Y., Perkins-Balding, D. & Thomson, J.G. Recombinase technology: applications and possibilities. Plant Cell Rep. 30, 267–285 (2011).
Callura, J.M., Cantor, C.R. & Collins, J.J. Genetic switchboard for synthetic biology applications. Proc. Natl. Acad. Sci. USA 109, 5850–5855 (2012).
Ham, T.S., Lee, S.K., Keasling, J.D. & Arkin, A.P. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS One 3, e2815 (2008).
Ham, T.S., Lee, S.K., Keasling, J.D. & Arkin, A.P. A tightly regulated inducible expression system utilizing the fim inversion recombination switch. Biotechnol. Bioeng. 94, 1–4 (2006).
Friedland, A.E. et al. Synthetic gene networks that count. Science 324, 1199–1202 (2009).
Bonnet, J., Subsoontorn, P. & Endy, D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. USA 109, 8884–8889 (2012).
Bonnet, J., Yin, P., Ortiz, M.E., Subsoontorn, P. & Endy, D. Amplifying genetic logic gates. Science 340, 599–603 (2013).
Groth, A.C. & Calos, M.P. Phage integrases: biology and applications. J. Mol. Biol. 335, 667–678 (2004).
Gordley, R.M., Gersbach, C.A. & Barbas, C.F. III. Synthesis of programmable integrases. Proc. Natl. Acad. Sci. USA 106, 5053–5058 (2009).
Daniel, R., Rubens, J.R., Sarpeshkar, R. & Lu, T.K. Synthetic analog computation in living cells. Nature 497, 619–623 (2013).
Gibson, D.G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Davis, J.H., Rubin, A.J. & Sauer, R.T. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 39, 1131–1141 (2011).
Lutz, R. & Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 (1997).
Young, J.W. et al. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nat. Protoc. 7, 80–88 (2012).
Locke, J.C.W. & Elowitz, M.B. Using movies to analyse gene circuit dynamics in single cells. Nat. Rev. Microbiol. 7, 383–392 (2009).
Isaacs, F.J. et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 22, 841–847 (2004).
Ghosh, P., Kim, A.I. & Hatfull, G.F. The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol. Cell 12, 1101–1111 (2003).
Ringrose, L., Chabanis, S., Angrand, P.O., Woodroofe, C. & Stewart, A.F. Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J. 18, 6630–6641 (1999).
Acknowledgements
We acknowledge G.F. Hatfull for the bxb1 gene and J.J. Collins for the riboregulator plasmids. We thank A.N. Billings and J. Rubens for help with the microscopy experiments and careful comments on the manuscript. This work was supported by the Defense Advanced Research Projects Agency (DARPA) and an Office of Naval Research Multidisciplinary University Research Initiative (MURI) grant. T.K.L. acknowledges support from the NIH New Innovator Award (1DP2OD008435).
Author information
Authors and Affiliations
Contributions
T.K.L. conceived of this study. All the experiments were implemented, constructed and performed by P.S. and J.Y. All authors analyzed the data, discussed the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
T.K.L. and P.S. have filed a provisional patent application (US patent application no. 14/105,994) on this work.
Rights and permissions
About this article
Cite this article
Siuti, P., Yazbek, J. & Lu, T. Engineering genetic circuits that compute and remember. Nat Protoc 9, 1292–1300 (2014). https://doi.org/10.1038/nprot.2014.089
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.089
This article is cited by
-
Digital circuits and neural networks based on acid-base chemistry implemented by robotic fluid handling
Nature Communications (2023)
-
Colloidal nanoelectronic state machines based on 2D materials for aerosolizable electronics
Nature Nanotechnology (2018)
-
An organic jelly made fractal logic gate with an infinite truth table
Scientific Reports (2015)
-
Construction of a genetic conditional learning system in Escherichia coli
Science China Information Sciences (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.