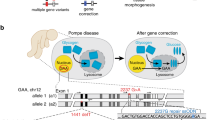
Abstract
I report here a detailed protocol for seamless genome editing using the piggyBac transposon in human pluripotent stem cells (hPSCs). Recent advances in custom endonucleases have enabled us to routinely perform genome editing in hPSCs. Conventional approaches use the Cre/loxP system that leaves behind residual sequences in the targeted genome. I used the piggyBac transposon to seamlessly remove a drug selection cassette and demonstrated safe genetic correction of a mutation causing α-1 antitrypsin deficiency in patient-derived hPSCs. An alternative approach to using the piggyBac transposon to correct mutations involves using single-stranded oligonucleotides, which is a faster process to complete. However, this experimental procedure is rather complicated and it may be hard to achieve homozygous modifications. In contrast, using the piggyBac transposon with drug selection–based enrichment of genetic modifications, as described here, is simple and can yield multiple correctly targeted clones, including homozygotes. Although two rounds of genetic manipulation are required to achieve homozygote modifications, the entire process takes ∼3 months to complete.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Capecchi, M.R. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6, 507–512 (2005).
Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F. & Cullin, C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3329–3330 (1993).
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002).
Winzeler, E.A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999).
Skarnes, W.C. et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011).
Thomson, J.A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Watanabe, K. et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686 (2007).
Hockemeyer, D. et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27, 851–857 (2009).
Hockemeyer, D. et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731–734 (2011).
Jasin, M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 12, 224–228 (1996).
Porteus, M.H. & Baltimore, D. Chimeric nucleases stimulate gene targeting in human cells. Science 300, 763 (2003).
Urnov, F.D. et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651 (2005).
Kim, Y.G. & Chandrasegaran, S. Chimeric restriction endonuclease. Proc. Natl. Acad. Sci. USA 91, 883–887 (1994).
Miller, J.C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Reyon, D. et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465 (2012).
Kim, Y. et al. A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 31, 251–258 (2013).
Schmid-Burgk, J.L., Schmidt, T., Kaiser, V., Honing, K. & Hornung, V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat. Biotechnol. 31, 76–81 (2013).
Ding, Q. et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 12, 238–251 (2013).
Doyon, Y. et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods 8, 74–79 (2011).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Ding, Q. et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12, 393–394 (2013).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 10.1038/nbt.2623 (2013).
Hsu, P.D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 10.1038/nbt.2647 (2013).
van der Weyden, L., Adams, D.J. & Bradley, A. Tools for targeted manipulation of the mouse genome. Physiol. Genomics 11, 133–164 (2002).
Meier, I.D. et al. Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression. FASEB J. 24, 1714–1724 (2010).
Yusa, K. et al. Targeted gene correction of α-antitrypsin deficiency in induced pluripotent stem cells. Nature 478, 391–394 (2011).
Tennessen, J.A. et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337, 64–69 (2012).
Conrad, D.F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010).
Maurano, M.T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Osafune, K. et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26, 313–315 (2008).
Kajiwara, M. et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 109, 12538–12543 (2012).
Chen, F. et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8, 753–755 (2011).
Soldner, F. et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 146, 318–331 (2011).
Friedel, R.H., Wurst, W., Wefers, B. & Kuhn, R. Generating conditional knockout mice. Methods Mol. Biol. 693, 205–231 (2011).
Rad, R. et al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science 330, 1104–1107 (2010).
Loonstra, A. et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA 98, 9209–9214 (2001).
Silver, D.P. & Livingston, D.M. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell 8, 233–243 (2001).
Vallier, L., Alexander, M. & Pedersen, R. Conditional gene expression in human embryonic stem cells. Stem Cells 25, 1490–1497 (2007).
Yusa, K., Zhou, L., Li, M.A., Bradley, A. & Craig, N.L. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. USA 108, 1531–1536 (2011).
Meng, X., Thibodeau-Beganny, S., Jiang, T., Joung, J.K. & Wolfe, S.A. Profiling the DNA-binding specificities of engineered Cys2His2 zinc finger domains using a rapid cell-based method. Nucleic Acids Res. 35, e81 (2007).
Yanover, C. & Bradley, P. Extensive protein and DNA backbone sampling improves structure-based specificity prediction for C2H2 zinc fingers. Nucleic Acids Res. 39, 4564–4576 (2011).
Zykovich, A., Korf, I. & Segal, D.J. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 37, e151 (2009).
Doyle, E.L. et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117–W122 (2012).
Pattanayak, V., Ramirez, C.L., Joung, J.K. & Liu, D.R. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 8, 765–770 (2011).
Gabriel, R. et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 29, 816–823 (2011).
Maeder, M.L., Thibodeau-Beganny, S., Sander, J.D., Voytas, D.F. & Joung, J.K. Oligomerized pool engineering (OPEN): an 'open-source' protocol for making customized zinc-finger arrays. Nat. Protoc. 4, 1471–1501 (2009).
Sanjana, N.E. et al. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7, 171–192 (2012).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Cary, L.C. et al. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172, 156–169 (1989).
Mitra, R., Fain-Thornton, J. & Craig, N.L. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 27, 1097–1109 (2008).
Wu, S.C. et al. piggyBac is a flexible and highly active transposon as compared to Sleeping Beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl. Acad. Sci. USA 103, 15008–15013 (2006).
Ding, S. et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005).
Fraser, M.J., Ciszczon, T., Elick, T. & Bauser, C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 5, 141–151 (1996).
Yusa, K., Rad, R., Takeda, J. & Bradley, A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nature Methods 6, 363–369 (2009).
Li, M.A. et al. The piggyBac transposon displays local and distant reintegration preferences and can cause mutations at noncanonical integration sites. Mol. Cell Biol. 33, 1317–1330 (2013).
Lacoste, A., Berenshteyn, F. & Brivanlou, A.H. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell 5, 332–342 (2009).
Rostovskaya, M. et al. Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res. 40, e150 (2012).
Beers, J. et al. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc. 7, 2029–2040 (2012).
Nandivada, H. et al. Fabrication of synthetic polymer coatings and their use in feeder-free culture of human embryonic stem cells. Nat. Protoc. 6, 1037–1043 (2011).
Braam, S.R. et al. Feeder-free culture of human embryonic stem cells in conditioned medium for efficient genetic modification. Nat. Protoc. 3, 1435–1443 (2008).
Lerou, P.H. et al. Derivation and maintenance of human embryonic stem cells from poor-quality in vitro fertilization embryos. Nat. Protoc. 3, 923–933 (2008).
Vallier, L. Serum-free and feeder-free culture conditions for human embryonic stem cells. Methods Mol. Biol. 690, 57–66 (2011).
Melkoumian, Z. et al. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat. Biotechnol. 28, 606–610 (2010).
Rodin, S. et al. Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nat. Biotechnol. 28, 611–615 (2010).
Villa-Diaz, L.G. et al. Synthetic polymer coatings for long-term growth of human embryonic stem cells. Nat. Biotechnol. 28, 581–583 (2010).
Schuldiner, M., Itskovitz-Eldor, J. & Benvenisty, N. Selective ablation of human embryonic stem cells expressing a 'suicide' gene. Stem Cells 21, 257–265 (2003).
Acknowledgements
I thank E.P. Tan for careful reading of the manuscript, and S.T. Rashid and members of the Allan Bradley laboratory (Sanger Institute, UK) and the Ludovic Vallier laboratory (University of Cambridge, UK) for discussion and for sharing their experience. This work was supported by Wellcome Trust (WT077187).
Author information
Authors and Affiliations
Contributions
K.Y. developed the protocol and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Yusa, K. Seamless genome editing in human pluripotent stem cells using custom endonuclease–based gene targeting and the piggyBac transposon. Nat Protoc 8, 2061–2078 (2013). https://doi.org/10.1038/nprot.2013.126
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.126
This article is cited by
-
WNT signalling control by KDM5C during development affects cognition
Nature (2024)
-
CRISPR Del/Rei: a simple, flexible, and efficient pipeline for scarless genome editing
Scientific Reports (2022)
-
MAPRE2 mutations result in altered human cranial neural crest migration, underlying craniofacial malformations in CSC-KT syndrome
Scientific Reports (2021)
-
Non-coding cis-element of Period2 is essential for maintaining organismal circadian behaviour and body temperature rhythmicity
Nature Communications (2019)
-
Updated summary of genome editing technology in human cultured cells linked to human genetics studies
Journal of Human Genetics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.