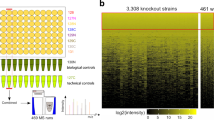
Abstract
Global protein expression profiling of various mutants or growth conditions is currently a major challenge in biology. Here we provide a protocol for a strategy that we recently developed that couples ORFeome-based (ORF denotes open reading frame) expression to reverse protein arrays; this approach accurately quantifies more than 99% of the predicted fission yeast proteins in various genetic backgrounds. The first stage of this two-stage protocol requires mass mating between any fertile fission yeast mutant of interest and the integrated fission yeast–tagged ORFeome followed by selection of recombinant haploids. The second stage of the protocol, called reverse protein arrays, involves simple large-scale extraction of total proteins, which are then spotted on nitrocellulose membranes for detection by quantitative dot blot. When handled manually, the entire protocol takes about 2 months. However, the process could easily be automated and should also be applicable to other organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Picotti, P., Bodenmiller, B., Mueller, L.N., Domon, B. & Aebersold, R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 (2009).
Uhlen, M. Affinity as a tool in life science. Biotechniques 44, 649–654 (2008).
Uhlen, M., Graslund, S. & Sundstrom, M. A pilot project to generate affinity reagents to human proteins. Nat. Methods 5, 854–855 (2008).
Colwill, K. & Graslund, S. A roadmap to generate renewable protein binders to the human proteome. Nat. Methods 8, 551–558 (2011).
Matsuyama, A. et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24, 841–847 (2006).
Kim, D.U. et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28, 617–623 (2010).
Spirek, M. et al. S. pombe genome deletion project: an update. Cell Cycle 9, 2399–2402 (2010).
Bauer, F. et al. Translational control of cell division by Elongator. Cell Rep. 1, 424–433 (2012).
Dewez, M. et al. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc. Natl. Acad. Sci. USA 105, 5459–5464 (2008).
Nugent, R.L. et al. Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation. BMC Genomics 11, 59 (2010).
Baryshnikova, A. et al. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol. 470, 145–179 (2010).
Coluccio, A.E., Rodriguez, R.K., Kernan, M.J. & Neiman, A.M. The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS ONE 3, e2873 (2008).
Maundrell, K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265, 10857–10864 (1990).
Bamps, S. et al. Mcs2 and a novel CAK subunit Pmh1 associate with Skp1 in fission yeast. Biochem. Biophys. Res. Commun. 325, 1424–1432 (2004).
Fersht, N., Hermand, D. & Nurse, P. Cdc18/CDC6 activates the Rad3-dependent checkpoint in the fission yeast. Nucleic Acids Res. 35, 5323–5337 (2007).
Coudreuse, D. et al. A gene-specific requirement of RNA polymerase II CTD phosphorylation for sexual differentiation in S. pombe. Curr. Biol. 20, 1053–1064 (2010).
Eisen, M.B., Spellman, P.T., Brown, P.O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868 (1998).
Acknowledgements
We thank the GEMO laboratory for discussions. This work was supported by a European Molecular Biology Organization (EMBO) short-term fellowship (58-2008), and grants FRFC 2.4510.10, Credit aux chercheurs 1.5.013.09 and MIS F.4523.11 to D.H.; F.B. is a Fonds de la Recherche en Industrie et Agriculture (FRIA) Research Fellow; D.H. is a Fonds National de la Recherche Scientifique (FNRS; Belgium) Research Associate.
Author information
Authors and Affiliations
Contributions
F.B. designed and performed the experiments, and wrote the manuscript; A.M. and M.Y. designed the reverse protein array strategy, and helped with its development; D.H. designed the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Comparison of α-tubulin levels between the wild type and the mutant strains. Total extracts of biological triplicates of wild type and mutant (Δelp3Δctu1) strains have been collected and separated by PAGE; transferred onto nitrocellulose membrane, incubated with anti-α–tubulin primary antibodies, secondary fluorescent antibodies and scanned to measure the fluorescent signal of each lane. The resulting raw and mean values are indicated. (PDF 319 kb)
Supplementary Figure 2
A representative membrane after hybridization and scanning. Total extracts of wild type ORFeome were spotted in quadruplicates onto nitrocellulose membrane, incubated with anti-α–tubulin and anti-His6 primary antibodies, secondary fluorescent antibodies and scanned to measure the fluorescent signal of each dot. As expected, the level of α-tubulin does not show strong variation from one strain to another while tagged ORFeome proteins display a much broader range of expression level. (PDF 1452 kb)
Rights and permissions
About this article
Cite this article
Bauer, F., Matsuyama, A., Yoshida, M. et al. Determining proteome-wide expression levels using reverse protein arrays in fission yeast. Nat Protoc 7, 1830–1835 (2012). https://doi.org/10.1038/nprot.2012.114
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.114
This article is cited by
-
Synchronized fission yeast meiosis using an ATP analog–sensitive Pat1 protein kinase
Nature Protocols (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.