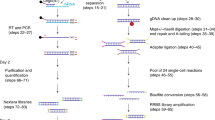
Abstract
DNA methylation is an epigenetic mark that has a crucial role in many biological processes. To understand the functional consequences of DNA methylation on phenotypic plasticity, a genome-wide analysis should be embraced. This in turn requires a technique that balances accuracy, genome coverage, resolution and cost, yet is low in DNA input in order to minimize the drain on precious samples. Methylated DNA immunoprecipitation-sequencing (MeDIP-seq) fulfils these criteria, combining MeDIP with massively parallel DNA sequencing. Here we report an improved protocol using 100-fold less genomic DNA than that commonly used. We show comparable results for specificity (>97%) and enrichment (>100-fold) over a wide range of DNA concentrations (5,000–50 ng) and demonstrate the utility of the protocol for the generation of methylomes from rare bone marrow cells using 160–300 ng of starting DNA. The protocol described here, i.e., DNA extraction to generation of MeDIP-seq library, can be completed within 3–5 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Goll, M.G. & Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 (2005).
Lister, R. et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011).
Hemberger, M. & Pedersen, R. Stem cells. Epigenome disruptors. Science 330, 598–599 (2010).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
He, Y.F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
Suzuki, M.M. & Bird, A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476 (2008).
Beck, S. & Rakyan, V.K. The methylome: approaches for global DNA methylation profiling. Trends Genet. 24, 231–237 (2008).
Laird, P.W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 11, 191–203 (2010).
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011).
Pastor, W.A. et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473, 394–397 (2011).
Down, T.A. et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 26, 779–785 (2008).
Beck, S. Taking the measure of the methylome. Nat. Biotechnol. 28, 1026–1028 (2010).
Borgel, J. et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42, 1093–1100 (2010).
Laurent, L. et al. Dynamic changes in the human methylome during differentiation. Genome Res. 20, 320–331 (2010).
Li, Y. et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 8, e1000533 (2010).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Smith, Z.D. et al. High-throughput bisulfite sequencing in mammalian genomes. Methods 48, 226–232 (2009).
Huang, Y. et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE 5, e8888 (2010).
Serre, D., Lee, B.H. & Ting, A.H. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 38, 391–399 (2010).
Rauch, T.A. & Pfeifer, G.P. DNA methylation profiling using the methylated-CpG island recovery assay (MIRA). Methods 52, 213–217 (2010).
Brinkman, A.B. et al. Whole-genome DNA methylation profiling using MethylCap-seq. Methods 52, 232–236 (2010).
Eckhardt, F. et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 38, 1378–1385 (2006).
Harris, R.A. et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 28, 1097–1105 (2010).
Butcher, L.M. & Beck, S. AutoMeDIP-seq: a high-throughput, whole genome, DNA methylation assay. Methods 52, 223–231 (2010).
Rakyan, V. et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res. 18, 1518–1529 (2008).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Chavez, L. et al. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 20, 1441–1450 (2010).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Acknowledgements
O.T. was supported by a PhD studentship from the UK Medical Research Council. S.S. was supported by the Boehringer Ingelheim Fonds, the Biotechnology and Biological Sciences Research Council (BBSRC) and Cambridge European Trust. W.R. is a Senior Investigator of the Wellcome Trust, and work in the Reik laboratory was supported by the BBSRC, Medical Research Council (MRC), EU Network of Excellence EpiGenesys, and Cellcentric. Work in the Beck laboratory was supported by the Wellcome Trust (084071), a Royal Society Wolfson Research Merit Award (WM100023), MRC (G1000411), the Engineering and Physical Sciences Research Council (EPSRC) (P14187), Innovative Medicines Initiative—Joint Undertaking (IMI-JU) OncoTrack (115234), and EU Seventh Framework Programme (EU-FP7) projects HEROIC (018883), EPIGENESYS (257082), IDEAL (259679), ITFoM (085602) and BLUEPRINT (282510).
Author information
Authors and Affiliations
Contributions
O.T., L.M.B. and S.B. conceived the study. S.S. and W.R. contributed data (early version of the protocol). D.P. contributed materials. O.T. and L.M.B. did the experiments and analyzed data. G.A.W. and T.M. did the bioinformatics analysis. O.T., L.M.B., G.A.W. and S.B. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Taiwo, O., Wilson, G., Morris, T. et al. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat Protoc 7, 617–636 (2012). https://doi.org/10.1038/nprot.2012.012
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.012
This article is cited by
-
SEEMLIS: a flexible semi-automated method for enrichment of methylated DNA from low-input samples
Clinical Epigenetics (2022)
-
The H3K27M mutation alters stem cell growth, epigenetic regulation, and differentiation potential
BMC Biology (2022)
-
Novel data archival system for multi-omics data of human exposure to harmful substances
Molecular & Cellular Toxicology (2022)
-
Dnmt1 has de novo activity targeted to transposable elements
Nature Structural & Molecular Biology (2021)
-
EpiMOLAS: an intuitive web-based framework for genome-wide DNA methylation analysis
BMC Genomics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.