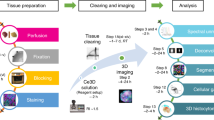
Abstract
In recent years, many mouse models have been developed to mark and trace the fate of adult cell populations using fluorescent proteins. High-resolution visualization of such fluorescent markers in their physiological setting is thus an important aspect of adult stem cell research. Here we describe a protocol to produce sections (150–200 μm) of near-native tissue with optimal tissue and cellular morphology by avoiding artifacts inherent in standard freezing or embedding procedures. The activity of genetically expressed fluorescent proteins is maintained, thereby enabling high-resolution three-dimensional (3D) reconstructions of fluorescent structures in virtually all types of tissues. The procedure allows immunofluorescence labeling of proteins to depths up to 50 μm, as well as a chemical 'Click-iT' reaction to detect DNA-intercalating analogs such as ethynyl deoxyuridine (EdU). Generation of near-native sections ready for imaging analysis takes approximately 2–3 h. Postsectioning processes, such as antibody labeling or EdU detection, take up to 10 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W.W. & Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994).
Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. 'Green mice' as a source of ubiquitous green cells. FEBS Lett. 407, 313–319 (1997).
Srinivas, S. et al. Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev. Genet. 24, 241–251 (1999).
Yoshimizu, T. et al. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 41, 675–684 (1999).
Hadjantonakis, A.K., Dickinson, M.E., Fraser, S.E. & Papaioannou, V.E. Technicolour transgenics: imaging tools for functional genomics in the mouse. Nat. Rev. Genet. 4, 613–625 (2003).
Morris, R.J. et al. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411–417 (2004).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Snippert, H.J. et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385–1389 (2010).
Tumbar, T. et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004).
Yoshida, S. et al. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev. Biol. 269, 447–458 (2004).
Montarras, D. et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science 309, 2064–2067 (2005).
Snippert, H.J. & Clevers, H. Tracking adult stem cells. EMBO Rep. 12, 113–122 (2011).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Mao, X., Fujiwara, Y., Chapdelaine, A., Yang, H. & Orkin, S.H. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood 97, 324–326 (2001).
Luche, H., Weber, O., Nageswara Rao, T., Blum, C. & Fehling, H.J. Faithful activation of an extra-bright red fluorescent protein in 'knock-in' Cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol. 37, 43–53 (2007).
Muzumdar, M.D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
De Gasperi, R. et al. The IRG mouse: a two-color fluorescent reporter for assessing Cre-mediated recombination and imaging complex cellular relationships in situ. Genesis 46, 308–317 (2008).
Yamamoto, M. et al. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis 47, 107–114 (2009).
Snippert, H.J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Dieguez-Hurtado, R. et al. A cre-reporter transgenic mouse expressing the far-red fluorescent protein katushka. Genesis 49, 36–45 (2011).
Desciak, E.B. & Maloney, M.E. Artifacts in frozen section preparation. Dermatol. Surg. 26, 500–504 (2000).
Snippert, H.J. et al. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology 136, 2187–2194 e2181 (2009).
Barker, N. et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Schepers, A.G., Vries, R., van den Born, M., van de Wetering, M. & Clevers, H. Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 30, 1104–1109 (2011).
Acknowledgements
We thank J. Kuipers and the Hubrecht Imaging Center for their support.
Author information
Authors and Affiliations
Contributions
H.J.S. and A.G.S. conducted experiments and wrote the manuscript, G.D. and P.D.S. provided materials, and H.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Video 1
Embedding tissue in LMA, Procedure Steps 4-7. (MOV 23985 kb)
Supplementary Video 2
Sectioning LMA-embedded tissue and mounting near-native sections, Procedure Steps 7-12, 14 and 15. (MOV 24883 kb)
Supplementary Table 1
Antibodies successfully used for staining near-native tissue sections. (XLSX 13 kb)
Rights and permissions
About this article
Cite this article
Snippert, H., Schepers, A., Delconte, G. et al. Slide preparation for single-cell–resolution imaging of fluorescent proteins in their three-dimensional near-native environment. Nat Protoc 6, 1221–1228 (2011). https://doi.org/10.1038/nprot.2011.365
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.365
This article is cited by
-
Advances and challenges in organ-on-chip technology: toward mimicking human physiology and disease in vitro
Medical & Biological Engineering & Computing (2024)
-
Tracing oncogene-driven remodelling of the intestinal stem cell niche
Nature (2021)
-
Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury
Nature (2018)
-
Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach
Nature Cell Biology (2017)
-
BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes
Nature Communications (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.