Abstract
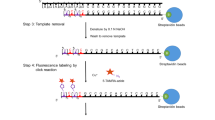
This protocol describes a homogeneous, convenient and sensitive DNA methylation detection method, using an optically amplifying cationic conjugated polymer (CCP, poly((1,4-phenylene)-2,7-[9,9-bis(6′-N,N,N-trimethyl ammonium)-hexyl fluorene] dibromide)). Genomic DNA from cancer cells is pretreated with a methylation-sensitive restriction endonuclease, followed by PCR amplification in the presence of fluorescein-labeled dNTP and Taq polymerase. The PCR only occurs for methylated DNA. DNA methylation of the gene sequence of interest is detected as a result of the fluorescence resonance energy transfer (FRET) between CCP and fluorescein that is incorporated into DNA. The methylated statuses of the p16, HPP1 and GALR2 promoters of five cancer cell lines (HT29, HepG2, A498, HL60 and M17) were assayed to provide an association study between the cancers and susceptibility genes, which shows great potential for early cancer diagnosis. This protocol simplifies previously available procedures by avoiding the need for primer labeling, isolation or purification steps, and sophisticated instruments. The assay takes about 20 h to obtain the methylated statuses of cancer cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Reik, W. & Walter, J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2, 21–32 (2001).
Robertson, K.D. & Wolffe, A.P. DNA methylation in health and disease. Nat. Rev. Genet. 1, 11–19 (2000).
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002).
Baylin, S.B. & Herman, J.G. DNA Alterations in Cancer: Genetic and Epigenetic Alterations 293–309 (Eaton Publishing, Natick, Massachusetts, 2000).
Antequera, F. & Bird, A. Number of CpG islands and genes in human and mouse. Proc. Natl. Acad. Sci. USA 90, 11995–11999 (1993).
Esteller, M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 62, 4519–4524 (2002).
Szyf, M. DNA Methylation and Cancer Therapy (Springer-Verlag, Berlin, 2005).
Feng, F., Wang, H., Han, L. & Wang, S. Fluorescent conjugated polyelectrolyte as an indicator for convenient detection of DNA methylation. J. Am. Chem. Soc. 130, 11338–11343 (2008).
Kuo, K.C., McCune, R.A., Gehrke, C.W., Midgett, R. & Ehrlich, M. Quantitative reversed-phase high performance liquid chromatographic determination of major and modified deoxyribonucleosides in DNA. Nucleic Acids Res. 8, 4763–4776 (1980).
Herman, J.G., Graff, J.R., Myohanen, S., Nelkin, B.D. & Baylin, S.B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 93, 9821–9826 (1996).
Song, G.A. et al. Mucinous carcinomas of the colorectum have distinct molecular genetic characteristics. Int. J. Oncol. 26, 745–750 (2005).
Costello, J.F. et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 24, 132–138 (2000).
Shiraishi, M. et al. Methyl-CpG binding domain column chromatography as a tool for the analysis of genomic DNA methylation. Anal. Biochem. 329, 1–10 (2004).
Hatada, I., Hayashizaki, Y., Hirotsune, S., Komatsubara, H. & Mukai, T. A genomic scanning method for higher organisms using restriction sites as landmarks. Proc. Natl. Acad. Sci. USA 88, 9523–9527 (1991).
Singer-Sam, J., LeBon, J.M., Tanguay, R.L. & Riggs, A.D. A quantitative HpaII-PCR assay to measure methylation of DNA from a small number of cells. Nucleic Acids Res. 18, 687 (1990).
Gonzalgo, M.L. & Jones, P.A. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res. 25, 2529–2531 (1997).
Xiong, Z. & Laird, P.W. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 25, 2532–2534 (1997).
Hayatsu, H. The bisulfite genomic sequencing used in the analysis of epigenetic states, a technique in the emerging environmental genotoxicology research. Mutat. Res. 659, 77–82 (2008).
Frommer, M. et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 89, 1827–1831 (1992).
Clark, S.J., Harrison, J., Paul, C.L. & Frommer, M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22, 2990–2997 (1994).
Dobrovic, A Molecular Diagnostics: For the Clinical Laboratorian 149–160 (Humana Press, Totowa, New Jersey, 2005).
Pfeifer, G.P., Steigerwald, S.D., Mueller, P.R., Wold, B. & Riggs, A.D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science 246, 810–813 (1989).
Gonzalgo, M.L. et al. Identification and characterization of differentially methylated regions of genomic DNA by methylation-sensitive arbitrarily primed PCR. Cancer Res. 57, 594–599 (1997).
El-Maarri, O., Herbiniaux, U., Walter, J. & Oldenburg, J. A rapid, quantitative, non-radioactive bisulfite-SNuPE- IP RP HPLC assay for methylation analysis at specific CpG sites. Nucleic Acids Res. 30, e25 (2002).
Tost, J., Schatz, P., Schuster, M., Berlin, K. & Gut, I.G. Analysis and accurate quantification of CpG methylation by MALDI mass spectrometry. Nucleic Acids Res. 31, e50 (2003).
Eads, C.A. et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 28, e32 (2000).
Thomas, S.W., Joly, G.D. & Swager, T.M. Chemical sensors based on amplifying fluorescent conjugated polymers. Chem. Rev. 107, 1339–1386 (2007).
Feng, F. et al. Fluorescent conjugated polyelectrolytes for biomacromolecule detection. Adv. Mater. 20, 2959–2964 (2008).
Gaylord, B.S., Heeger, A.J. & Bazan, G.C. DNA hybridization detection with water-soluble conjugated polymers and chromophore-labeled single-stranded DNA. J. Am. Chem. Soc. 125, 896–900 (2003).
Pu, K.-Y. & Liu, B. Optimizing the cationic conjugated polymer-sensitized fluorescent signal of dye labeled oligonucleotide for biosensor applications. Biosens. Bioelectron. 24, 1067–1073 (2009).
Förster, T. Intramolecular energy migration and fluorescence. Ann. Phys. 2, 55–75 (1948).
Duan, X., Li, Z., He, F. & Wang, S. A sensitive and homogeneous SNP detection using cationic conjugated polymers. J. Am. Chem. Soc. 129, 4154–4155 (2007).
Duan, X., Liu, L. & Wang, S. Homogeneous and one-step fluorescent allele-specific PCR for SNP genotyping assays using conjugated polyelectrolytes. Biosens. Bioelectron. 24, 2095–2099 (2009).
Ratge, D., Scheiblhuber, B., Landt, O., Berg, J. & Knabbe, C. Two-round rapid-cycle RT-PCR in single closed capillaries increases the sensitivity of HCV RNA detection and avoids amplicon carry-over. J. Clin. Virol. 24, 161–172 (2002).
Kwok, S. & Higuchi, R. Avoiding false positives with PCR. Nature 339, 237–238 (1989).
Duan, X. et al. Single-nucleotide polymorphism (SNP) genotyping using cationic conjugated polymers in homogeneous solution. Nat. Protoc. 4, 984–991 (2009).
Sambrook, J. & Russell, D.W. Molecular Cloning: A Laboratory Manual 3rd ed. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001).
Acknowledgements
We are grateful to the National Natural Science Foundation of China (Nos. 90913014 and 20725308) and to the Major Research Plan of China (No. 2006CB932102).
Author information
Authors and Affiliations
Contributions
F.F. conducted experiments and data analysis, and wrote the paper; L.L. conducted experiments and data analysis; S.W. designed experiments, performed data analysis and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Feng, F., Liu, L. & Wang, S. Fluorescent conjugated polymer-based FRET technique for detection of DNA methylation of cancer cells. Nat Protoc 5, 1255–1264 (2010). https://doi.org/10.1038/nprot.2010.79
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.79
This article is cited by
-
A novel fluorescent biosensor based on dendritic DNA nanostructure in combination with ligase reaction for ultrasensitive detection of DNA methylation
Journal of Nanobiotechnology (2019)
-
Technical advances in global DNA methylation analysis in human cancers
Journal of Biological Engineering (2017)
-
Convenient, Sensitive and High-Throughput Method for Screening Botanic Origin
Scientific Reports (2014)
-
Fluorescence resonance energy transfer microscopy as demonstrated by measuring the activation of the serine/threonine kinase Akt
Nature Protocols (2013)
-
Formation and Characterization of Stable Fluorescent Complexes Between Neutral Conjugated Polymers and Cyclodextrins
Journal of Fluorescence (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.