Abstract
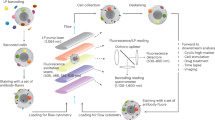
Aptamers that target a specific cell subpopulation within composite mixtures represent invaluable tools in biomedical research and in the development of cell-specific therapeutics. Here we describe a detailed protocol for a modular and generally applicable scheme to select aptamers that target the subpopulations of cells in which you are interested. A fluorescence-activated cell-sorting device is used to simultaneously differentiate and separate those subpopulations of cells having bound and unbound aptamers. There are fewer false positives when using this approach in comparison with other cell-selection approaches in which unspecific binding of nucleic acids to cells with reduced membrane integrity or their unselective uptake by dead cells occurs more often. The protocol provides a state-of-the-art approach for identifying aptamers that selectively target virtually any cell type under investigation. As an example, we provide the step-by-step protocol targeting CD19+ Burkitt's lymphoma cells, starting from the pre-SELEX (systematic evolution of ligands by exponential amplification) measurements to establish suitable SELEX conditions and ending at completion of the SELEX procedure, which reveals the enriched single-stranded DNA library.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Neri, D., Pini, A. & Nissim, A. Antibodies from phage display libraries as immunochemical reagents. Methods Mol. Biol. 80, 475–500 (1998).
Marlind, J. et al. Antibody-mediated delivery of interleukin-2 to the stroma of breast cancer strongly enhances the potency of chemotherapy. Clin. Cancer Res. 14, 6515–6524 (2008).
Schliemann, C. & Neri, D. Antibody-based targeting of the tumor vasculature. Biochim. Biophys. Acta. 1776, 175–192 (2007).
Neri, D., Petrul, H. & Roncucci, G. Engineering recombinant antibodies for immunotherapy. Cell Biophys. 27, 47–61 (1995).
Gafner, V., Trachsel, E. & Neri, D. An engineered antibody-interleukin-12 fusion protein with enhanced tumor vascular targeting properties. Int. J. Cancer 119, 2205–2212 (2006).
Jäger, S. & Famulok, M. Generation and enzymatic amplification of high-density functionalized DNA double strands. Angew. Chem. Int. Ed. Engl. 43, 3337–3340 (2004).
Jäger, S. et al. A versatile toolbox for variable DNA functionalization at high density. J. Am. Chem. Soc. 127, 15071–15082 (2005).
Thum, O., Jäger, S. & Famulok, M. Functionalized DNA: a new replicable biopolymer. Angew. Chem. Int. Ed. Engl. 40, 3990–3993 (2001).
Verma, S. & Eckstein, F. Modified oligonucleotides: synthesis and strategy for users. Annu. Rev. Biochem. 67, 99–134 (1998).
Mayer, G. The chemical biology of aptamers. Angew. Chem. Int. Ed. Engl. 48, 2672–2689 (2009).
Shamah, S.M., Healy, J.M. & Cload, S.T. Complex target SELEX. Acc. Chem. Res. 41, 130–138 (2008).
Famulok, M., Hartig, J.S. & Mayer, G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 107, 3715–3743 (2007).
Famulok, M. & Verma, S. In vivo-applied functional RNAs as tools in proteomics and genomics research. Trends Biotechnol. 20, 462–466 (2002).
Cerchia, L., Giangrande, P.H., McNamara, J.O. & de Franciscis, V. Cell-specific aptamers for targeted therapies. Methods Mol. Biol. 535, 59–78 (2009).
Guo, K.T. et al. A new technique for the isolation and surface immobilization of mesenchymal stem cells from whole bone marrow using high-specific DNA aptamers. Stem Cells 24, 2220–2231 (2006).
Pastor, F., Kolonias, D., Giangrande, P.H. & Gilboa, E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 465, 227–230 (2010).
Ellington, A.D. & Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Hermann, T. & Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 287, 820–825 (2000).
Huang, D.B. et al. Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer. Proc. Natl Acad. Sci. USA 100, 9268–9273 (2003).
Jaeger, J., Restle, T. & Steitz, T.A. The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor. EMBO J. 17, 4535–4542 (1998).
Kettenberger, H. et al. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat. Struct. Mol. Biol. 13, 44–48 (2006).
Sassanfar, M. & Szostak, J.W. An RNA motif that binds ATP. Nature 364, 550–553 (1993).
Burgstaller, P. & Famulok, M. Isolation of RNA aptamers for biological cofactors by in vitro selection. Angew. Chem. Int. Ed. 33, 1084–1087 (1994).
Famulok, M. Molecular recognition of amino-acids by RNA-aptamers—an L-citrulline binding RNA motif and its evolution into an L-arginine binder. J. Am. Chem. Soc. 116, 1698–1706 (1994).
Nastasijevic, B., Becker, N.A., Wurster, S.E. & Maher, L.J. III . Sequence-specific binding of DNA and RNA to immobilized nickel ions. Biochem. Biophys. Res. Commun. 366, 420–425 (2008).
Proske, D., Hofliger, M., Soll, R.M., Beck-Sickinger, A.G. & Famulok, M. A Y2 receptor mimetic aptamer directed against neuropeptide Y. J. Biol. Chem. 277, 11416–11422 (2002).
Rentmeister, A., Bill, A., Wahle, T., Walter, J. & Famulok, M. RNA aptamers selectively modulate protein recruitment to the cytoplasmic domain of beta-secretase BACE1 in vitro. RNA 12, 1650–1660 (2006).
Zichi, D., Eaton, B., Singer, B. & Gold, L. Proteomics and diagnostics: let's get specific, again. Curr. Opin. Chem. Biol. 12, 78–85 (2008).
Mayer, G., Raddatz, M.S., Grunwald, J.D. & Famulok, M. RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch. Angew. Chem. Int. Ed. Engl. 46, 557–560 (2007).
Rentmeister, A., Mayer, G., Kuhn, N. & Famulok, M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 35, 3713–3722 (2007).
Rentmeister, A., Mayer, G., Kuhn, N. & Famulok, M. Secondary structures and functional requirements for thiM riboswitches from Desulfovibrio vulgaris, Erwinia carotovora and Rhodobacter spheroides. Biol. Chem. 389, 127–134 (2008).
Lunse, C.E. et al. An aptamer targeting the apical-loop domain modulates pri-miRNA processing. Angew. Chem. Int. Ed. Engl. 49, 4674–4677 (2010).
Mayer, G. & Hover, T. In vitro selection of ssDNA aptamers using biotinylated target proteins. Methods Mol. Biol. 535, 19–32 (2009).
Hicke, B.J. et al. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 276, 48644–48654 (2001).
Li, N., Larson, T., Nguyen, H.H., Sokolov, K.V. & Ellington, A.D. Directed evolution of gold nanoparticle delivery to cells. Chem. Commun. (Camb.) 46, 392–394 (2010).
Cerchia, L. et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 3, e123 (2005).
Blank, M., Weinschenk, T., Priemer, M. & Schluesener, H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 276, 16464–16468 (2001).
Sefah, K., Shangguan, D., Xiong, X., O'Donoghue, M.B. & Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 5, 1169–1185 (2010).
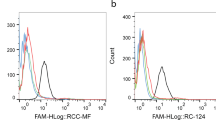
Raddatz, M.S. et al. Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angew. Chem. Int. Ed. Engl. 47, 5190–5193 (2008).
Li, N. et al. Technical and biological issues relevant to cell typing with aptamers. J. Proteome. Res. 8, 2438–2448 (2009).
Lupold, S.E., Hicke, B.J., Lin, Y. & Coffey, D.S. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 62, 4029–4033 (2002).
Proske, D. et al. Prion-protein-specific aptamer reduces PrPSc formation. Chembiochem 3, 717–725 (2002).
Maecker, H.T. & Trotter, J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry A 69, 1037–1042 (2006).
Perfetto, S.P., Ambrozak, D., Nguyen, R., Chattopadhyay, P. & Roederer, M. Quality assurance for polychromatic flow cytometry. Nat. Protoc. 1, 1522–1530 (2006).
Ahmed, M.S. & Mayer, G. Evolution of specific RNA motifs derived from pan-protein interacting precursors. Bioorg. Med. Chem. Lett. 20, 3793–3796 (2010).
Mayer, G. et al. From selection to caged aptamers: identification of light-dependent ssDNA aptamers targeting cytohesin. Bioorg. Med. Chem. Lett. 19, 6561–6564 (2009).
Mayer, G. et al. An RNA molecule that specifically inhibits G-protein-coupled receptor kinase 2 in vitro. RNA 14, 524–534 (2008).
Paul, A., Avci-Adali, M., Ziemer, G. & Wendel, H. P. Streptavidin-coated magnetic beads for DNA strand separation implicate a multitude of problems during cell-SELEX. Oligonucleotides 19, 243–254 (2009).
Mallikaratchy, P., Stahelin, R.V., Cao, Z., Cho, W. & Tan, W. Selection of DNA ligands for protein kinase C-delta. Chem. Commun. (Camb.) 2006, 3229–3231 (2006).
Zueva, E., Rubio, L.I., Duconge, F. & Tavitian, B. Metastasis-focused cell-based SELEX generates aptamers inhibiting cell migration and invasion. Int. J. Cancer published online, doi:10.1002/ijc.25401 (19 April 2010).
Acknowledgements
This research was supported by research grants from the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 704, the Graduiertenkolleg GRK 804 and the Fonds der Chemischen Industrie. We thank N. Krämer for excellent technical assistance and J. Müller for support during the preparation of peripheral blood mononuclear cells from whole blood.
Author information
Authors and Affiliations
Contributions
G.M. and M.-S.L.A., along with M.F., performed and designed most of the included studies. E.E. and A.D. provided experimental input in setting up the FACS experiments and performed some of them to establish optimal conditions, assisted by M.-S.L.A. and G.M. P.A.K. provided conceptual input. M.F. and G.M. supervised the research project and assisted in the experimental design. All authors discussed the experimental results. M.F. and G.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Mayer, G., Ahmed, MS., Dolf, A. et al. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat Protoc 5, 1993–2004 (2010). https://doi.org/10.1038/nprot.2010.163
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.163
This article is cited by
-
Aptamer-based Membrane Protein Analysis and Molecular Diagnostics
Chemical Research in Chinese Universities (2024)
-
Selection of DNA Aptamers Against Neisseria gonorrhoeae Causing Sexually Transmitted Infection (STI)
Molecular Biotechnology (2023)
-
Preparation, applications, and challenges of functional DNA nanomaterials
Nano Research (2023)
-
Research progress of whole-cell-SELEX selection and the application of cell-targeting aptamer
Molecular Biology Reports (2022)
-
A comparative analysis of cell surface targeting aptamers
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.