Abstract
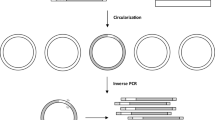
Brachypodium distachyon is emerging as a new model system for bridging research into temperate cereal crops, such as wheat and barley, and for promoting research in novel biomass grasses. Here, we provide an adapter ligation PCR protocol that allows the large-scale characterization of T-DNA insertions into the genome of Brachypodium. The procedure enables the retrieval and mapping of the regions flanking the right and left borders (RB and LB) of the T-DNA inserts and consists of five steps: extraction and restriction digest of genomic DNA; ligation of an adapter to the genomic DNA; PCR amplification of the regions flanking the T-DNA insert(s) using primers specific to the adapter and the T-DNA; sequencing of the PCR products; and identification of the flanking sequence tags (FSTs) characterizing the T-DNA inserts. Analyzing the regions flanking both the LB and RB of the T-DNA inserts significantly improves FST retrieval and the frequency of mutant lines for which at least one FST can be identified. It takes approximately 16 or 10 d for a single person to analyze 96 T-DNA lines using individual or batch procedures, respectively.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Hirochika, H. et al. Rice mutant resources for gene discovery. Plant Mol. Biol. 54, 325–334 (2004).
Meinke, D., Muralla, R., Sweeney, C. & Dickerman, A. Identifying essential genes in Arabidopsis thaliana . Trends Plant Sci. 13, 483–491 (2008).
Garvin, D.F. et al. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci. 48, S69–S84 (2008).
Opanowicz, M., Vain, P., Draper, J., Parker, D. & Doonan, J.H. Brachypodium distachyon: making hay with a wild grass. Trends Plant Sci. 13, 172–177 (2008).
Draper, J. et al. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127, 1539–1555 (2001).
Griffiths, S. et al. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439, 749–752 (2006).
Springer, P.S. Gene traps: tools for plant development and genomics. Plant Cell 12, 1007–1020 (2000).
Jung, K.H., An, G.H. & Ronald, P.C. Towards a better bowl of rice: assigning function to tens of thousands of rice genes. Nat. Rev. Genet. 9, 91–101 (2008).
Vain, P. et al. Agrobacterium-mediated transformation of the temperate grass Brachypodium distachyon (genotype Bd21) for T-DNA insertional mutagenesis. Plant Biotechnol. J. 6, 236–245 (2008).
Gelvin, S.B. Agobacterium-mediated plant transformation: the biology behind the 'gene-Jockeying' tool. Microbiol. Mol. Biol. Rev. 67, 16–37 (2003).
Martineau, B., Voelker, T.A. & Sanders, R.A. On defining T-DNA. Plant Cell 6, 1032–1033 (1994).
Ulker, B. et al. T-DNA-mediated transfer of Agrobacterium tumefaciens chromosomal DNA into plants. Nat. Biotechnol. 26, 1015–1017 (2008).
Afolabi, A.S., Worland, B., Snape, J.W. & Vain, P. A large-scale study of rice plants transformed with different T-DNAs provides new insights into locus composition and T-DNA linkage configurations. Theor. Appl. Genet. 109, 815–826 (2004).
Vain, P., Afolabi, A.S., Worland, B. & Snape, J.W. Transgene behaviour in populations of rice plants transformed using a new dual binary vector system: pGreen/pSoup. Theor. Appl. Genet. 107, 210–217 (2003).
Thole, V., Worland, B., Snape, J.W. & Vain, P. The pCLEAN dual binary vector system for Agrobacterium-mediated plant transformation. Plant Physiol. 145, 1211–1219 (2007).
Murray, F., Brettell, R., Matthews, P., Bishop, D. & Jacobsen, J. Comparison of Agrobacterium-mediated transformation of four barley cultivars using the GFP and GUS reporter genes. Plant Cell Rep. 22, 397–402 (2004).
Spertini, D., Beliveau, C. & Bellemare, G. Screening of transgenic plants by amplification of unknown genomic DNA flanking T-DNA. Biotechniques 27, 308–314 (1999).
Strizhov, N. et al. High-throughput generation of sequence indexes from T-DNA mutagenized Arabidopsis thaliana lines. Biotechniques 35, 1164–1168 (2003).
Alonso, J.M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana . Science 301, 653–657 (2003).
Jeon, J.S. et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22, 561–570 (2000).
Sallaud, C. et al. Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor. Appl. Genet. 106, 1396–1408 (2003).
Jeong, D.H. et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 45, 123–132 (2006).
O'Malley, R.C., Alonso, J.M., Kim, C.J., Leisse, T.J. & Ecker, J.R. An adapter ligation-mediated PCR method for high-throughput mapping of T-DNA inserts in the Arabidopsis genome. Nat. Protoc. 2, 2910–2917 (2007).
Acknowledgements
This study was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) and through a Short-Term Marie Curie EST fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Thole, V., Alves, S., Worland, B. et al. A protocol for efficiently retrieving and characterizing flanking sequence tags (FSTs) in Brachypodium distachyon T-DNA insertional mutants. Nat Protoc 4, 650–661 (2009). https://doi.org/10.1038/nprot.2009.32
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2009.32
This article is cited by
-
Introduction of Multiple Genes via Agrobacterium-Mediated Co-Transformation into Soybean and Confirmation of Insertion Using PCR
Journal of Plant Biology (2024)
-
Genetic modification strategies for enhancing plant resilience to abiotic stresses in the context of climate change
Functional & Integrative Genomics (2023)
-
The APAF1_C/WD40 repeat domain-encoding gene from the sea lettuce Ulva mutabilis sheds light on the evolution of NB-ARC domain-containing proteins in green plants
Planta (2022)
-
Development of event-specific detection method for identification of insect resistant NIBGE-1601 cotton harboring double gene Cry1Ac-Cry2Ab construct
Scientific Reports (2021)
-
Cyclic Digestion and Ligation-Mediated PCR Used for Flanking Sequence Walking
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.