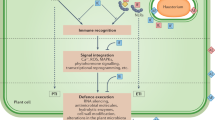
Abstract
A pathogenic model in which both the pathogen and its host are amenable to genetic manipulation can greatly facilitate the understanding of bacterial pathogenesis. Plants are genetically tractable and can be used as experimental models for human microbial pathogenesis. We present protocols for both lettuce and Arabidopsis leaf infection models using the opportunistic human bacterial pathogen, Pseudomonas aeruginosa. The lettuce model allows for high-throughput qualitative analysis of virulence and is suitable for screening large numbers of bacterial strains, whereas the Arabidopsis model provides a quantitative approach and permits the tracking of bacterial cell proliferation in planta. The lettuce model takes ∼24 h including bacterial growth using store-bought lettuce, and the Arabidopsis model takes 4–6 weeks to grow the plants and a similar time as with lettuce to infect the plants. Both models are monitored for up to 5 d post-infection. These methodologies can and have been used to identify novel and critical P. aeruginosa pathogenicity agents, as virulence factors are often conserved across phylogeny.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Rahme, L.G. et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902 (1995).
Nordmann, P., Naas, T., Fortineau, N. & Poirel, L. Superbugs in the coming new decade; multidrug resistance and prospects for treatment of Staphylococcus aureus, Enterococcus spp. and Pseudomonas aeruginosa in 2010. Curr. Opin. Microbiol. 10, 436–440 (2007).
Ausubel, F.M. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979 (2005).
Rahme, L.G. et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902 (1995).
Apidianakis, Y. et al. Involvement of skeletal muscle gene regulatory network in susceptibility to wound infection following trauma. PLoS ONE 2, e1356 (2007).
Apidianakis, Y. et al. Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc. Natl. Acad. Sci. USA 102, 2573–2578 (2005).
Mahajan-Miklos, S., Tan, M.-W., Rahme, L.G. & Ausubel, F.M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell 96, 47–56 (1999).
Cosson, P. et al. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184, 3027–3033 (2002).
Mahajan-Miklos, S., Rahme, L.G. & Ausubel, F.M. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol. Microbiol. 37, 981–988 (2000).
Lau, G.W. et al. The Drosophila melanogaster toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa . Infect. Immun. 71, 4059–4066 (2003).
Hoffmann, J.A. The immune response of Drosophila . Nature 426, 33–38 (2003).
Hoffmann, J.A. et al. Phylogenetic perspectives in innate immunity. Science 284, 1313–1318 (1999).
Shan, L., He, P. & Sheen, J. Intercepting host MAPK signaling cascades by bacterial type III effectors. Cell Host Microbe 1, 167–174 (2007).
Apostol, I., Heinstein, P.F. & Low, P.S. Rapid stimulation of an oxidative burst during elicitation of cultured plant cells: role in defense and signal transduction. Plant Physiol. 90, 109–116 (1989).
Doke, N. NADPH-dependent O2-generation in membrane fractions isolated from wounded potato tubers inoculated with Phytophthora infestans . Physiol. Plant Pathol. 27, 311–322 (1985).
Ohashi, Y., Brickman, J.M., Furman, E., Middleton, B. & Carey, M. Modulating the potency of an activator in a yeast in vitro transcription system. Mol. Cell Biol. 14, 2731–2739 (1994).
Sutherland, M.W. The generation of oxygen radicals during host plant responses to infection. Physiol. Mol. Plant Pathol. 39, 79–94 (1991).
Hunter, P. Common defences. EMBO Rep. 6, 504–507 (2005).
Broekaert, W.F., Terras, F.R., Cammue, B.P. & Osborn, R.W. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol. 108, 1353–1358 (1995).
Ganz, T. & Lehrer, R.I. Defensins. Curr. Opin. Immunol. 6, 584–589 (1994).
van Baarlen, P., van Belkum, A. & Thomma, B.P.H.J. Disease induction by human microbial pathogens in plant-model systems: potential, problems and prospects. Drug Discov. Today 12, 167–173 (2007).
Jha, A.K., Bais, H.P. & Vivanco, J.M. Enterococcus faecalis mammalian virulence-related factors exhibit potent pathogenicity in the Arabidopsis thaliana plant model. Infect Immun. 73, 464–475 (2005).
Prithiviraj, B., Bais, H.P., Jha, A.K. & Vivanco, J.M. Staphylococcus aureus pathogenicity on Arabidopsis thaliana is mediated either by a direct effect of salicylic acid on the pathogen or by SA-dependent, NPR1-independent host responses. Plant J. 42, 417–432 (2005).
Rahme, L.G. et al. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 94, 13245–13250 (1997).
Rahme, L.G. et al. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97, 8815–8821 (2000).
Dong, X., Mindrinos, M., Davis, K.R. & Ausubel, F.M. Induction of Arabidopsis defense genes by virulent and Avirulent Pseudomonas syringae strains and by a cloned Avirulence gene. Plant Cell 3, 61–72 (1991).
Katagiri, F., Thilmony, R. & He, S.Y. The Arabidopsis thaliana–Pseudomonas syringae interaction. The Arabidopsis Book, Rockville, MD: American Society of Plant Biologists. DOI: 10.1199/tab.0039 (2002).
Plotnikova, J.M., Rahme, L.G. & Ausubel, F.M. Pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa PA14 in Arabidopsis . Plant Physiol. 124, 1766–1774 (2000).
Walker, T.S. et al. Pseudomonas aeruginosa–plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 134, 320–331 (2004).
Baldini, R.L., Lau, G.W. & Rahme, L.G. Use of plant and insect hosts to model bacterial pathogenesis. Methods Enzymol. 358, 3–13 (2002).
Silo-Suh, L., Suh, S.-J., Sokol, P.A. & Ohman, D.E. A simple alfalfa seedling infection model for Pseudomonas aeruginosa strains associated with cystic fibrosis shows AlgT (sigma-22) and RhlR contribute to pathogenesis. Proc. Natl. Acad. Sci. USA 99, 15699–15704 (2002).
Attila, C. et al. Pseudomonas aeruginosa PAO1 virulence factors and poplar tree response in the rhizosphere. Microbial Biotechnol. 1, 17–29 (2008).
Kaplan, E. & Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 53, 457–481 562–563 (1958).
Jander, G., Rahme, L.G. & Ausubel, F.M. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182, 3843–3845 (2000).
Miyata, S., Casey, M., Frank, D.W., Ausubel, F.M. & Drenkard, E. Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect. Immun. 71, 2404–2413 (2003).
Fauvarque, M.O. et al. Role and activation of type III secretion system genes in Pseudomonas aeruginosa-induced Drosophila killing. Microb. Pathog. 32, 287–295 (2002).
Liberati, N.T. et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103, 2833–2838 (2006).
Jacobs, M.A. et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa . Proc. Natl. Acad. Sci. USA 100, 14339–14344 (2003).
Acknowledgements
This work was partially supported by the research grant R01AI063433. M.S. was supported by the Shriners Research Fellowship no. 8506. Thank you to Jenifer Bush for growing and maintaining our Arabidopsis plants.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Starkey, M., Rahme, L. Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nat Protoc 4, 117–124 (2009). https://doi.org/10.1038/nprot.2008.224
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2008.224
This article is cited by
-
Microgravity and evasion of plant innate immunity by human bacterial pathogens
npj Microgravity (2023)
-
Characterization of three new plant growth-promoting microbes and effects of the interkingdom interactions on plant growth and disease prevention
Plant Cell Reports (2023)
-
Comparative Effects of Azospirillum brasilense Sp245 and Pseudomonas aeruginosa PAO1 Lipopolysaccharides on Wheat Seedling Growth and Peroxidase Activity
Journal of Plant Growth Regulation (2021)
-
An immune receptor complex evolved in soybean to perceive a polymorphic bacterial flagellin
Nature Communications (2020)
-
Transgenic tobacco expressing Medicago sativa Defensin (Msdef1) confers resistance to various phyto-pathogens
The Nucleus (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.