Abstract
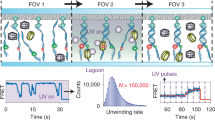
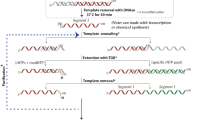
Hydroxyl radical (·OH) footprinting provides comprehensive site-specific quantitative information about the structural changes associated with macromolecular folding, interactions and ligand binding. 'Fast Fenton' footprinting is a laboratory-based method for time-resolved ·OH footprinting capable of millisecond time resolution readily applicable to DNA and RNA. This protocol utilizes inexpensive chemical reagents (H2O2, Fe(NH4)2(SO4)2, EDTA, thiourea or ethanol) and widely available quench-flow mixers to reveal transient, often short-lived, intermediate states of complex biochemical processes. We describe a protocol developed to study RNA folding that can be readily tailored to particular applications. Once familiar with quench-flow mixer operation and its calibration, nucleic acid labeling and the conduct of a dose–response experiment, a single kinetic experiment of 30 time points takes about 1 h to perform. Sample processing and separation of the ·OH reaction products takes several hours. Data analysis can take 45 min to several weeks depending on the depth of analysis conducted.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others

References
Galas, D.J. & Schmitz, A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5, 3157–3170 (1978).
Latham, J.A. & Cech, T.R. Defining the inside and outside of a catalytic RNA molecule. Science 245, 276–282 (1989).
Petri, V. & Brenowitz, M. Quantitative nucleic acids footprinting: thermodynamic and kinetic approaches. Curr. Opin. Biotechnol. 8, 36–44 (1997).
Buxton, G.V., Greenstock, C.L., Helman, W.P. & Ross, A.B. Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms and hydroxyl radicals in aqueous solution. J. Phys. Chem. Ref. Data 17, 513–886 (1988).
Tullius, T.D. & Dombroski, B.A. Hydroxyl radical 'footprinting': high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc. Natl. Acad. Sci. USA 83, 5469–5473 (1986).
Hsieh, M. & Brenowitz, M. Quantitative kinetics footprinting of protein-DNA association reactions. Methods Enzymol. 274, 478–492 (1996).
Fenton, H.J.H. The oxidation of tartaric acid in presence of iron. J. Chem. Soc. 65, 899–910 (1894).
Shcherbakova, I., Mitra, S., Beer, R.H. & Brenowitz, M. Fast Fenton footprinting: a laboratory-based method for the time-resolved analysis of DNA, RNA and proteins. Nucleic Acids Res. 34, e48 (2006).
Shcherbakova, I., Mitra, S., Beer, R.H. & Brenowitz, M. Following molecular transitions with single residue spatial and millisecond time resolution. in Methods in Cell Biology, Vol. 84 (eds. Correia, J.J. & Detrich, H.W.) 589–615 (Elsevier/Academic Press, New York, 2008).
Tullius, T.D. & Dombroski, B.A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science 230, 679–681 (1985).
Sclavi, B., Sullivan, M., Chance, M.R., Brenowitz, M. & Woodson, S.A. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science 279, 1940–1943 (1998).
Ralston, C.Y. et al. Time-resolved synchrotron X-ray footprinting and its application to RNA folding. Methods Enzymol. 317, 353–368 (2000).
Brenowitz, M., Chance, M.R., Dhavan, G. & Takamoto, K. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical 'footprinting'. Curr. Opin. Struct. Biol. 12, 648–653 (2002).
Aye, T.T., Low, T.Y. & Sze, S.K. Nanosecond laser-induced photochemical oxidation method for protein surface mapping with mass spectrometry. Anal. Chem. 77, 5814–5822 (2005).
Hambly, D.M. & Gross, M.L. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J. Am. Soc. Mass Spectrom. 16, 2057–2063 (2005).
Heyduk, T., Heyduk, E., Severinov, K., Tang, H. & Ebright, R.H. Determinants of RNA polymerase alpha subunit for interaction with beta, beta′, and sigma subunits: hydroxyl-radical protein footprinting. Proc. Natl. Acad. Sci. USA 93, 10162–10166 (1996).
Sharp, J.S., Becker, J.M. & Hettich, R.L. Protein surface mapping by chemical oxidation: structural analysis by mass spectrometry. Anal. Biochem. 313, 216–225 (2003).
Takamoto, K. & Chance, M.R. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu. Rev. Biophys. Biomol. Struct. 35, 251–276 (2006).
Sclavi, B. et al. Real-time characterization of intermediates in the pathway to open complex formation by Escherichia coli RNA polymerase at the T7A1 promoter. Proc. Natl. Acad. Sci. USA 102, 4706–4711 (2005).
Zaug, A.J., Grosshans, C.A. & Cech, T.R. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry 27, 8924–8931 (1988).
Huang, Z. & Szostak, J.W. A simple method for 3′-labeling of RNA. Nucleic Acids Res. 24, 4360–4361 (1996).
Shcherbakova, I., Gupta, S., Chance, M.R. & Brenowitz, M. Monovalent ion-mediated folding of the Tetrahymena thermophila ribozyme. J. Mol. Biol. 342, 1431–1442 (2004).
Henegariu, O., Bray-Ward, P. & Ward, D.C. Custom fluorescent-nucleotide synthesis as an alternative method for nucleic acid labeling. Nat. Biotechnol. 18, 345–348 (2000).
Nguyenle, T., Laurberg, M., Brenowitz, M. & Noller, H.F. Following the dynamics of changes in solvent accessibility of 16 S and 23 S rRNA during ribosomal subunit association using synchrotron-generated hydroxyl radicals. J. Mol. Biol. 359, 1235–1248 (2006).
Adilakshmi, T., Ramaswamy, P. & Woodson, S.A. Protein-independent folding pathway of the 16S rRNA 5′ domain. J. Mol. Biol. 351, 508–519 (2005).
Brenowitz, M., Senear, D.F., Shea, M.A. & Ackers, G.K. Quantitative DNase footprint titration: a method for studying protein–DNA interactions. Methods Enzymol. 130, 132–181 (1986).
Sclavi, B., Woodson, S., Sullivan, M., Chance, M. & Brenowitz, M. Following the folding of RNA with time-resolved synchrotron X-ray footprinting. Methods Enzymol. 295, 379–402 (1998).
Das, R., Laederach, A., Pearlman, S.M., Herschlag, D. & Altman, R.B. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11, 344–354 (2005).
Laederach, A., Shcherbakova, I., Liang, M.P., Brenowitz, M. & Altman, R.B. Local kinetic measures of macromolecular structure reveal partitioning among multiple parallel pathways from the earliest steps in the folding of a large RNA molecule. J. Mol. Biol. 358, 1179–1190 (2006).
Sharp, J.S., Becker, J.M. & Hettich, R.L. Analysis of protein solvent accessible surfaces by photochemical oxidation and mass spectrometry. Anal. Chem. 76, 672–683 (2004).
Tijerina, P., Mohr, S. & Russell, R. DMS footprinting of structured RNAs and RNA–protein complexes. Nat. Protoc. 2, 2608–2623 (2007).
Sclavi, B., Woodson, S., Sullivan, M., Chance, M.R. & Brenowitz, M. Time-resolved synchrotron X-ray 'footprinting', a new approach to the study of nucleic acid structure and function: application to protein–DNA interactions and RNA folding. J. Mol. Biol. 266, 144–159 (1997).
Laederach, A., Shcherbakova, I., Jonikas, M.A., Altman, R.B. & Brenowitz, M. Distinct contribution of electrostatics, initial conformational ensemble, and macromolecular stability in RNA folding. Proc. Natl. Acad. Sci. USA 104, 7045–7050 (2007).
Acknowledgements
The writing of this article was supported by National Institutes of Health grant PO1–GM066275 from the Institute of General Medical Sciences. We thank Jörg Schlatterer, Somdeb Mitra, Alain Laederach, Rick Russell and Yaqi Wan for critically reading the manuscript and confirming the details of the presented protocol.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Shcherbakova, I., Brenowitz, M. Monitoring structural changes in nucleic acids with single residue spatial and millisecond time resolution by quantitative hydroxyl radical footprinting. Nat Protoc 3, 288–302 (2008). https://doi.org/10.1038/nprot.2007.533
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.533
This article is cited by
-
Millisecond-order rapid micromixing with non-equilibrium electrokinetic phenomena
Microfluidics and Nanofluidics (2012)
-
Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution
Nature Protocols (2009)
-
Footprinting protein–DNA complexes using the hydroxyl radical
Nature Protocols (2008)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.