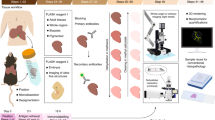
Abstract
This protocol details methods for the isolation of oocyte nuclear envelopes (NEs) from the African clawed toad Xenopus laevis, immunogold labeling of component proteins and subsequent visualization by field-emission scanning electron microscopy (FESEM) and transmission electron microscopy (TEM). This procedure involves the initial removal of the ovaries from mature female X. laevis, the dissection of individual oocytes, then the manual isolation of the giant nucleus and subsequent preparation for high-resolution visualization. Unlike light microscopy, and its derivative technologies, electron microscopy enables 3–5 nm resolution of nuclear structures, thereby giving unrivalled opportunities for investigation and immunological characterization in situ of nuclear structures and their structural associations. There are a number of stages where samples can be stored, although we recommend that this protocol take no longer than 2 d. Samples processed for FESEM can be stored for weeks under vacuum, allowing considerable time for image acquisition.
Similar content being viewed by others
Introduction
Biological material derived from the amphibian Xenopus laevis has been used for decades as an experimental system for the elucidation and characterization of complex cellular events and structures. Study of nuclear structures and dynamics often involves the visualization of protein localization and distribution by conventional direct or indirect immunofluorescence. However, these studies, by their nature, give only limited information on the structural context in which these proteins and complexes are found. Therefore, our laboratory has focused on optimizing protocols for the isolation and high-resolution visualization of oocyte nuclei, nuclei formed in vitro and nuclei derived from both yeast and tissue culture cells1,2,3,4,5,6,7,8,9,10,11,12. Detailed protocols for the visualization of nuclei formed in cell-free extracts of Xenopus eggs and tissue culture cell nuclei13,14 can be found in separate manuscripts available at http://www.natureprotocols.com.
The application of field-emission scanning electron microscopy (FESEM) and transmission electron microscopy (TEM) for high-resolution visualization of biological specimens has revealed the 3D structure of macromolecular assemblies such as nuclear pore complexes (NPCs) that mediate the essential, highly dynamic process of nucleo–cytoplasmic transport.
The giant nucleus of Xenopus oocytes (often referred to as the germinal vesicle) is a valuable source of material for visualization and study of nuclear structures at high resolutions, specifically by FESEM and TEM. FESEM yields highly detailed visual information about the surface topology of the sample and is especially useful for determining and examining the structural microenvironment in which specific macromolecular complexes are found. In contrast, TEM involves the sectioning of samples that allows the visualization of internal structures often obscured and thus unavailable for FESEM imaging. However, such sample sectioning generates only a 2D image of the sample, although 3D images of the sample can be generated from serial sectioning and image reconstitution using appropriate computer software (e.g., Reconstruc15). The nuclei of Xenopus oocytes can be manually dissected and are a rich source of NPCs with approximately 50 million NPCs per nucleus, in contrast to approximately 5,000 in the nuclear envelope (NE) of a somatic cell nucleus. These oocyte nuclei have proved to be important in determining the component structures of the NPC and their associations with the NE and underlying nuclear lamina4,5,6.
Interestingly, comparative 3D cryo-EM mapping studies of the yeast and vertebrate NPC has revealed a striking conservation in structure3, thereby supporting the cross-species validity of the ultrastructural data generated by high-resolution visualization of Xenopus oocyte NPCs. The use of immunogold labeling of defined proteins and protein domains with polyclonal or monoclonal antibodies has yielded a wealth of information on the location of individual proteins within the NPC, their topology and the modular NPC structures they are part of (e.g., the localization of nup153 (ref. 9), nup214 and nup358/RanBP2 (ref. 10)). FESEM allows the visualization of only surface morphologies; however, due to the relatively large size of oocyte nuclei (approximately 4–600 μm in diameter), it has been possible to visualize the nuclear interior. Kiseleva et al.8 revealed that NPCs are connected by an intra-nuclear actin-dependent filament network that also interacts with Cajal bodies and, to a lesser extent, chromatin. The large amphibian nucleus contains relatively little peripheral chromatin compared with somatic nuclei and, therefore, is ideal for visual investigation of internal filamentous structures that would otherwise be obscured by chromosomes. In addition, oocyte nuclei are highly active for transcription and nucleocytoplasmic transport and are relatively easy to use in microinjection procedures for intranuclear addition of exogenous compounds or DNA or RNA constructs.
Here, we detail methods for the visualization of amphibian oocyte nuclei from the removal of the oocytes from the source animal, through the manual dissection of the giant nuclei and then the processing of these nuclei for SEM and TEM with immunogold labeling of component proteins.
Materials
Reagents
-
Acetone (VWR International)
-
Argon cylinder, high purity greater than 99.999% (Air Liquide)
Caution
Cylinders should be fitted by suitably trained staff.
-
Ringers A (see REAGENT SETUP)
-
Ringers B (see REAGENT SETUP)
-
Amphibian Ringers (see REAGENT SETUP)
-
Calcium chloride (Sigma-Aldrich, cat. no. C3306) (see REAGENT SETUP)
-
CO2 cylinder, liquid with dip tube (less than 5 p.p.m. water) (Air Liquide)
Caution
Cylinders should be fitted by suitably trained staff.
-
Ethanol, 30, 50, 70 and 95% solutions in dH2O, and 100% (VWR International, cat. no. 101077Y)
-
Glutaraldehyde, EM grade (Fluka, cat. no. 49631) (see REAGENT SETUP)
Caution
Toxic, observe Material Safety Data Sheet (MSDS) advice.
-
HEPES (Sigma-Aldrich, cat. no. H7523) (see REAGENT SETUP)
-
Osmium tetroxide, 1% in dH2O (Agar Scientific, cat. no. R1017)
Caution
Toxic, observe MSDS advice.
-
Potassium chloride (Sigma-Aldrich, cat. no. P9541) (see REAGENT SETUP)
-
Potassium hydroxide, 10 M in dH2O (VWR International, cat. no.105021)
Caution
Corrosive, observe MSDS advice.
-
Sodium carbonate (Sigma-Aldrich, cat. no. S7795) (see REAGENT SETUP)
-
Tannic acid (TAAB laboratories, cat. no. T046) (see REAGENT SETUP)
-
Uranyl acetate, 1 and 2% in dH2O (Agar Scientific, cat. no. R1260A). Can be stored for several weeks in the fridge if protected from light
Caution
Toxic, observe MSDS advice.
-
X. laevis
Caution
General animal husbandry and use must be in accordance with relevant guidelines and regulations.
-
BSA (Sigma-Aldrich, cat. no. A7030) (see REAGENT SETUP)
-
Fish skin gelatin (Sigma-Aldrich, cat. no. G7765) (see REAGENT SETUP)
-
Gly (Sigma-Aldrich, cat. no. G7403) (see REAGENT SETUP)
-
Paraformaldehyde (TAAB laboratories, cat. no. P001/1) (see REAGENT SETUP)
Caution
Harmful, observe MSDS advice.
-
Primary antibody, as appropriate to research (commercially available or custom made)
-
Secondary antibody 10-nm gold conjugate EM grade, as appropriate for primary antibody (GE Healthcare)
-
Agar 100 resin kit (Agar Scientific, cat. no. R1031) (see REAGENT SETUP)
Caution
Observe MSDS advice.
-
Lead nitrate (Fluka, cat. no. 15334) (see REAGENT SETUP)
Caution
Toxic, observe MSDS advice.
-
Citric acid trisodium salt (Sigma-Aldrich, cat. no. C3674) (see REAGENT SETUP)
-
Sodium hydroxide (VWR International, cat. no. 106495) (see REAGENT SETUP)
Caution
Corrosive, observe MSDS advice.
-
Fixative (see REAGENT SETUP)
Equipment
-
35-mm Petri dishes biological, e.g., BD Falcon (Biotrace International)
-
35-mm Petri dishes tissue culture, e.g., BD Falcon (Biotrace International)
-
Basket stem assembly (Leica Microsystems, cat. no. 16709248)
-
Bunsen or Butane burner (VWR International)
Caution
Follow equipment instructions.
-
Coating unit with chromium target capable of depositing 2–3 nm layers of chromium with a grain size of less than 0.4 nm. (e.g., Edwards BOC) (see EQUIPMENT SETUP)
-
Critical point dryer (e.g., BAL-TEC, Liechtenstein) (see EQUIPMENT SETUP)
Caution
Follow equipment instructions.
-
Dissecting microscope, variable zoom with a black base plate and two flexible light sources for incident illumination (e.g., Leica Microsystems)
-
Dissecting needle (VWR International)
-
Fine glass needles (see EQUIPMENT SETUP)
-
Disposable baskets, small four divisions (Leica Microsystems, cat. no. 16709298)
-
Dumont tweezers 3 Dumostar (Agar Scientific)
-
Dumont tweezers 5 Dumostar, 2 pairs (Agar Scientific)
-
Glass Pasteur pipettes (VWR International) (see EQUIPMENT SETUP)
-
High-resolution scanning electron microscope
-
Pegwood sticks (Agar Scientific)
-
Rubber pipette filler for Pasteur pipettes. (VWR International)
-
Silicon chips 5 × 5 mm2 (Agar Scientific) (see EQUIPMENT SETUP)
-
Vacuum desiccator capable of achieving 1 × 10−2 mbar
-
Writing diamond (Agar Scientific)
-
90-mm plastic Petri dishes (VWR International)
-
Backscattered electron detector fitted to SEM
-
Filter paper Whatman grade no. 50 (VWR International)
-
Nescofilm (VWR International)
-
Copper grids 200 mesh hexagonal (Agar Scientific)
-
Dumont tweezers 7 Dumostar (Agar Scientific)
-
Embedding oven (TAAB laboratories equipment)
-
Glass beakers 50 ml (VWR International)
-
Grid box (Agar Scientific)
-
Plastic disposable pipettes 2 ml (VWR)
-
Polyethylene graduated containers with caps (Agar Scientific)
-
TEM
-
Ultramicrotome (Leica)
Reagent setup
-
5:1/HEPES buffer 83 mM KCl, 17 mM NaCl, 10 mM HEPES in dH2O, pH 7.5. Prepare 1 M solutions of KCl, NaCl and HEPES and add in appropriate volumes with dH2O for final concentrations. Use 10 M KOH to pH the HEPES buffer.
-
Ringers A (5 × stock) 55 mM NaCl, 9.5 mM KCl, 5.5 mM CaCl2.
-
Ringers B (5 × stock) 2% NaHCO3.
-
Amphibian Ringers 111 mM NaCl, 1.9 mM KCl, 1.1 mM CaCl2, 2.4 mM NaHCO3. Add 200 ml Ringers A (5 ×) + 750 ml dH2O +10 ml Ringers B (5 ×) + 40 ml dH2O.
-
Fixative 2% Glutaraldehyde, 0.2% tannic acid in 100 mM HEPES, pH 7.4 freshly prepared and filtered using 0.4 μm syringe filter.
-
BSA 1% in 5:1/HEPES. Prepare fresh on day of use.
-
Fish skin gelatin 1% in 5:1/HEPES.
Critical
Prepare fresh on day of use.
-
Fixative 2% Paraformaldehyde, 0.01% glutaraldehyde in 5:1/HEPES pH 7.5. Dissolve paraformaldehyde in buffer by heating up to 80 °C with agitation, allow to cool; add glutaraldehyde.
Critical
Prepare fresh on day of use.
-
Gly 0.1 M in 5:1/HEPES.
Critical
Prepare fresh on day of use.
-
Agar 100 resin Prepare fresh on day of use from reagents supplied in the kit. Agar 100, dodecinyl succinic anhydride (DDSA), methyl nadic anhydride (MNA), 2.4.6 tri(dimethylethylaminoethyl) phenol (DMP30) in a ratio of 19.2:10.4:10.4:0.5, respectively. Mix carefully using pegwood sticks avoiding the introduction of air bubbles. Allow to stand for 15 min before use. Prepare dilutions of one part resin to two parts ethanol and two parts resin to one part ethanol.
-
Lead citrate Weigh lead nitrate 1.33 g, and citric acid trisodium salt 1.76 g. Place in empty 50-ml flask, add 30 ml water, shake for 1 min, allow to stand for 30 min. Add 8 ml of 1 M sodium hydroxide then add water to make total 50 ml. Alternatively, add 1–2 pellets of sodium hydroxide and shake until clear then top up to 50 ml. It is not necessary to allow to stand for 30 min with second method. Store at 4 °C for as long as the solution remains clear.
Equipment setup
-
Coating unit Attach argon cylinder and set up to sputter chromium. Further details can be found in ref. 1.
-
Critical point dryer Attach liquid CO2 cylinder. Follow manufacturer's instructions for use.
-
Fine glass needles Prepare from a Pasteur pipette drawn out over a Bunsen flame. The optimal diameter is 0.025 mm (Fig. 1)
-
Fine glass pipette Prepare from a Pasteur pipette drawn out over a Bunsen flame. The optimum diameter is 0.5 mm (Fig. 2; top)
-
Silicon chips Number chips using the writing diamond. Clean by dipping in a beaker of acetone, and drying with tissue. Store in a covered Petri dish to protect from dust.
Procedure
Isolation of nuclear envelope
-
1
Remove ovary from Xenopus16.
Critical Step
Observe national legislation for handling animals.
Pause point
Store in amphibian Ringers at 4 °C for up to 1 week or 20 °C for up to 1–2 d.
-
2
Wash oocytes in fresh amphibian Ringers, remove small clusters of approximately 20 oocytes and transfer to 5:1/HEPES buffer. (Fig. 3) All incubations should be done at room temperature (approximately 20 °C) using sufficient volumes to prevent the oocytes from drying out.
-
3
Transfer the oocytes to a 35-mm Petri dish containing fresh 5:1/HEPES buffer and observe under a dissecting microscope with fibre-optic illumination. Carry out all subsequent manipulations under the binoculars. Select oocytes at the required stage of development, stage 5 or 6, 0.8–1.0 mm diameter (see Fig. 4). Use no. 5 tweezers to manipulate the oocytes (Supplementary Video 1 online).
-
4
Pierce the oocyte with a dissecting needle, avoiding the center of the dark hemisphere where the nucleus is located. The nucleus, visible as a transparent sphere, will be extruded with the cytoplasm (Supplementary Video 1).
-
5
Clean the nucleus of excess cytoplasm by pipetting carefully up and down in a fine glass pipette that has an internal diameter slightly larger than the nucleus (Supplementary Video 1).
Critical Step
Do not allow any air bubbles to touch the nucleus, or else it will lyse and disappear.
-
6
For FESEM, carefully transfer the nucleus to a silicon chip (shiny side) in fresh 5:1/HEPES buffer in an adjacent Petri dish (Supplementary Video 1). For TEM, transfer the nucleus to 35-mm tissue culture Petri dishes.
-
7
Allow the nucleus to settle. It will attach to the surface of the chip (or Petri dish) within a few seconds. Pipette the buffer against the nucleus very gently to remove any remaining cytoplasm. Carefully break open the nucleus using fine glass needles and continue to pipette the buffer against the NE to remove the nuclear contents. The envelope will attach to the chip (or Petri dish). Use the fine glass needles to further spread and affix the envelope (Supplementary Video 1).
Critical Step
With practice, it will be possible to attach more than one NE to each chip, but one per chip is advisable for beginners.
Critical Step
Ensure that there is sufficient buffer above the NE, and cover the dish with its lid to prevent the envelope from drying out.
Pause point
Store NEs at room temperature (approximately 20 °C) until a sufficient number has been processed.
-
8
Repeat Steps 4–7 until the required number of NEs have been obtained. At least two envelopes per subsequent treatment up to a maximum of 16 chips would be a reasonable number to process at one time. If immunolabeling is required, follow the protocol outlined in Box 1 (Fig. 5) before proceeding to Step 9. Otherwise, continue immediately with Step 9.
-
9
Transfer the chip with NE (quickly without air drying) to a 0.5-ml tube containing fixative (2% glutaraldehyde, 0.2% tannic acid in 100 mM HEPES, pH 7.4). NEs attached to Petri dishes should be fixed in situ and subsequent steps carried out within the Petri dish. It is helpful to mark the position of the envelope by circling the area with the writing diamond. Fix for a minimum of 10 min at room temperature (approximately 20 °C). This fixative has been optimized for visualization of NPC ultrastructure by FESEM and also works well for TEM.
Critical Step
For the following steps, use no. 3 tweezers to transfer chips manually into a Petri dish containing the appropriate reagent. For envelopes attached to Petri dishes, washes are added directly. Keep the NE upward. Do not allow to dry out at any point.
Pause point
Store in fixative at 4 °C for up to 1 week.
-
10
Wash briefly in dH2O.
-
11
Post fix in aqueous 1% osmium tetroxide for 10 min. Osmium preserves lipid and phospholipid content and also enhances contrast in the specimen.
-
12
Wash briefly in dH2O.
-
13
Stain for 10 min in 1% aqueous uranyl acetate. This stains nucleic acids and proteins.
-
14
To process samples for SEM, follow option A. To process for TEM, follow option B.
-
A
Processing for FESEM
-
i
Dehydrate through the following series of 2 min ethanol washes: 30, 50, 70, 95 (2 ×) and 100% (3 ×).
-
ii
Transfer to critical point drying apparatus containing ethanol as intermediate reagent. Use a suitable holding device. A Leica basket stem assembly with small baskets is suitable for the Baltec CPD. (It may be necessary to shorten the stem slightly to fit in the chamber.) Critical point dry from high purity (less than 5 p.p.m. water) liquid CO2.
Pause point
Samples can be stored under vacuum for up to 1 week.
-
iii
Coat samples with 3-nm chromium.
Pause point
Samples can be stored under vacuum for several months.
-
iv
Visualize by high-resolution SEM (Fig. 6a–f).
Figure 6: Field-emission scanning electron microscopy (FESEM) imaging of the oocyte nuclear envelope. (a) Low-power scanning electron microscopy (SEM) of whole oocyte nuclear envelope spread on Si chip. cs = cytoplasmic surface, ns = nucleoplasmic surface. (b, scale bar = 300 nm) Higher magnification of overlapping area. (c, scale bar = 200 nm) High magnification image of the cytoplasmic face of the NE. (d, scale bar = 50 nm) High magnification of single NE basket. (e, f) Basket labeled with 10 nm colloidal gold conjugated to nucleoporin-specific antibodies (combined secondary electron and backscattered electron imaging).
Pause point
Store samples under vacuum for optimal preservation. They should last for several months and can be recoated with chromium if required.
-
i
-
B
Processing for TEM
-
i
Dehydrate samples with one part resin to two parts ethanol for 1 h.
-
ii
Replace with two parts resin to one part ethanol for 1 h.
-
iii
Replace with 100% resin for 1 h.
-
iv
Replace with fresh 100% resin for 4 h or overnight.
-
v
Replace with fresh 100% resin and polymerize in the embedding oven at 60 °C for 48 h.
-
vi
Remove from oven and allow to cool.
Pause point
Store at room temperature (approximately 20 °C) indefinitely.
-
vii
Locate the NE in the resin block and trim and cut sections 60–70 nm thickness using the ultramicrotome. Sections can be cut parallel to or perpendicular to the plane of the embedded resin and hence the envelope.
-
viii
Collect sections on 200-mesh hexagonal copper grids. Use Dumont tweezers 7 Dumostar to manipulate the grids.
Pause point
Store dry grids in grid box indefinitely.
-
ix
Contrast stain by immersing the grids in drops of lead citrate for 5 min.
-
x
Wash by immersion in three beakers of dH2O, ten dips in each beaker.
-
xi
Remove excess dH2O with filter paper (number 50).
Pause point
Store dry grid in grid box indefinitely.
-
xii
Observe in the TEM (Fig. 7).
Figure 7: TEM images of nuclear pore complexes.
-
i
-
A
Troubleshooting
Troubleshooting advice can be found in Table 1.
Timing
Isolation of ovary: Step 1, 1–2 h
Preparation of NEs: Steps 2–7, 2–6 h (depending on number of envelopes required)
Immunolabeling (optional): Step 8, 4 h or overnight (Box 1)
Fixation and washes: Steps 9–13, 40 min
Processing for FESEM: Step 14A(i–ii), 40 min; Step 14A(iii) approximately 1 h (depending on the coating system used and time required to achieve correct vacuum); Step 14A(iv) image acquisition depends on the number of samples
Processing for TEM: Step 14B(i–iv), 8–24 h; Step 14B(v), 48 h; Step 14B(vi–xi) depends on the number of samples; Step 14B(xii), image acquisition, depends on number of samples.
Anticipated results
It is critical to ensure that the starting material for this protocol is of the highest standard, as this will have the single largest influence on the quality of images that can be captured by either FESEM or TEM. Therefore, it is essential to ensure that animals are kept appropriately. Figure 6 shows several representative images of an isolated and 'spread' oocyte nucleus, indicating the high density of NPCs within the NE and the macromolecular structural components of both the nucleoplasmic and cytoplasmic faces of the NPC. When immunogold labeling specific proteins and protein domains, the secondary electron image (e.g., Fig. 6d–f) is acquired simultaneously and is in exact register with the backscatter image. Consequently, it is straightforward to superimpose the location of the gold particles (via imaging software) as shown in Figure 6e and f.
When visualizing gold-labeled samples by TEM (Fig. 7), it is important to determine the plane of sectioning to aid orientation and interpretation of the images generated.
Note: Supplementary information is available via the HTML version of this article.
References
Allen, T.D. et al. Three dimensional surface structure of the nucleus. Methods Cell Biol. 53, 125–138 (1998).
Bagley, S., Goldberg, M.W., Cronshaw, J.M., Rutherford, S. & Allen, T.D. The nuclear pore complex. J. Cell Sci. 113, 3885–3886 (2000).
Drummond, S.P., Rutherford, S.A., Sanderson, H.S. & Allen, T.D. High resolution analysis of mammalian nuclear structure throughout the cell cycle: implications for nuclear pore complex assembly during interphase and mitosis. Can. J. Physiol. Pharmacol. 84, 423–430 (2006).
Goldberg, M.W. & Allen, T.D. High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J. Cell Biol. 119, 1429–1440 (1992).
Goldberg, M.W. & Allen, T.D. The nuclear pore complex: three-dimensional surface structure revealed by field emission, in-lens scanning electron microscopy, with underlying structure uncovered by proteolysis. J. Cell Sci. 106 (Pt 1): 261–274 (1993).
Goldberg, M.W. & Allen, T.D. Structural and functional organization of the nuclear envelope. Curr. Opin. Cell Biol. 7, 301–309 (1995).
Kiseleva, E. et al. Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J. Struct. Biol. 145, 272–288 (2004).
Kiseleva, E. et al. Actin- and protein-4.1-containing filaments link nuclear pore complexes to subnuclear organelles in Xenopus oocyte nuclei. J. Cell Sci. 117 (Pt 12): 2481–2490 (2004).
Walther, T.C. et al. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20, 5703–5714 (2001).
Walther, T.C. et al. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158, 63–77 (2002).
Walther, T.C. et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113, 195–206 (2003).
Zhang, C., Goldberg, M.W., Moore, W.J., Allen, T.D. & Clarke, P.R. Concentration of Ran on chromatin induces decondensation, nuclear envelope formation and nuclear pore complex assembly. Eur. J. Cell Biol. 81, 623–633 (2002).
Allen, T.D. et al. Generation of cell-free extracts of Xenopus eggs and demembranated sperm chromatin for the assembly and isolation of in vitro-formed nuclei for western blotting and scanning electron microscopy (SEM). Nat. Protoc. 2, 1173–1177 (2007).
Allen, T.D. et al. Visualisation of the nucleus and nuclear envelope in situ by SEM in tissue culture cells. Nat. Protoc. 2, 1180–1184 (2007).
Fiala, J.C. Reconstruct: a free editor for serial section microscopy. J. Microsc. 218 (Pt 1): 52–61 (2005).
Smith, L.D., Xu, W. & Varnold, R.L. Oogenesis and oocyte isolation. Methods Cell Biol. 36, 45–60 (1991).
Acknowledgements
T.D.A., S.A.R., S.M., H.S.S. and S.P.D. would like to acknowledge the support of Cancer Research (CR) UK. E.K. and M.W.G. acknowledge the support of The Wellcome Trust, and F.G. similarly acknowledges the Medical Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Video 1
Isolation of the Nucleus from the Oocytes of Xenopus laevis (MOV 13572 kb)
Rights and permissions
About this article
Cite this article
Allen, T., Rutherford, S., Murray, S. et al. A protocol for isolating Xenopus oocyte nuclear envelope for visualization and characterization by scanning electron microscopy (SEM) or transmission electron microscopy (TEM). Nat Protoc 2, 1166–1172 (2007). https://doi.org/10.1038/nprot.2007.137
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.137
This article is cited by
-
Embryonic and adult isoforms of XLAP2 form microdomains associated with chromatin and the nuclear envelope
Cell and Tissue Research (2011)
-
Preparation of cells for assessing ultrastructural localization of nanoparticles with transmission electron microscopy
Nature Protocols (2010)
-
A protocol for isolation and visualization of yeast nuclei by scanning electron microscopy (SEM)
Nature Protocols (2007)
-
Generation of cell-free extracts of Xenopus eggs and demembranated sperm chromatin for the assembly and isolation of in vitro–formed nuclei for Western blotting and scanning electron microscopy (SEM)
Nature Protocols (2007)
-
Visualization of the nucleus and nuclear envelope in situ by SEM in tissue culture cells
Nature Protocols (2007)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.