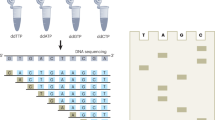
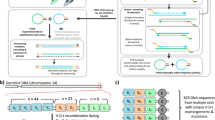
Abstract
The accurate analysis of genetic variation has major implications in many areas of biomedical research, including the identification of infectious agents (such as parasites), the diagnosis of infections, and the detection of unknown or known disease-causing mutations. Mutation scanning methods, including PCR-coupled single-strand conformation polymorphism (SSCP), have significant advantages over many other nucleic acid techniques for the accurate analysis of allelic and mutational sequence variation. The present protocol describes the SSCP method of analysis, including all steps from the small-scale isolation of genomic DNA and PCR amplification of target sequences, through to the gel-based separation of amplicons and scanning for mutations by SSCP (either by the analysis of radiolabeled amplicons in mutation detection enhancement (MDE) gels or by non-isotopic SSCP using precast GMA gels). The subsequent sequence analysis of polymorphic bands isolated from gels is also detailed. The SSCP protocol can readily detect point mutations for amplicon sizes of up to 450–500 bp, and usually takes 1–2 days to carry out. This user-friendly, low-cost, potentially high-throughput platform has demonstrated the utility to study a wide range of pathogens and diseases, and has the potential to be applied to any gene of any organism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gasser, R.B. Mutation scanning methods for the analysis of parasite genes. Int. J. Parasitol. 27, 1449–1463 (1997).
Cotton, R.G.H. Mutation Detection (Oxford University Press, Oxford, 1997).
Gasser, R.B. PCR-based technology in veterinary parasitology. Vet. Parasitol. 84, 229–258 (1999).
Orita, M., Suzuki, Y., Sekiya, T. & Hayashi, K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics 5, 874–879 (1989).
Hayashi, K. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Meth. Appl. 1, 34–38 (1991).
Hayashi, K., Kukita, Y., Inazuka, M. & Tahira, T. Single strand conformation polymorphism analysis. in Mutation Detection: A Practical Approach (eds. Cotton, R.G.H., Edkins, E. & Forrest, S.) 7–24 (Oxford University Press, Oxford, 1998).
Myers, R.M., Hedrick, L. & Hayashi, K. Detection of mutations. in Genome Analysis: A Laboratory Manual vol. 2 (eds. Green, E., Birren, B., Klapholz, S., Myer, R. & Hieter, P.) 287–384 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1998).
Dong, Y. & Zhu, H. Single-strand conformational polymorphism analysis: basic principles and routine practice. Methods Mol. Med. 108, 149–157 (2005).
Gasser, R.B. Molecular tools – advances, opportunities and prospects. Vet. Parasitol. 36, 69–89 (2006).
Gasser, R.B., Chilton, N.B., Hoste, H. & Beveridge, I. Rapid sequencing of rDNA from single worms and eggs of parasitic helminths. Nucleic Acids Res. 21, 2525–2526 (1993).
Gasser, R.B., Zhu, X.Q. & Chilton, N.B. The value of mutation scanning approaches for detecting genetic variation: implications for studying intestinal nematodes of humans. in World Class Parasites vol. 2 The Geohelminths: Ascaris, Trichuris and Hookworm (eds. Holland, C.V. & Kennedy, M.W.; Series eds. Black, S. & Seed, J.R.) 219–233 (Kluwer Academic Press, Boston, 2002).
Gasser, R., Nansen, P. & Guldberg, P. Fingerprinting sequence variation in ribosomal DNA of parasites by DGGE. Mol. Cell. Probes 10, 99–105 (1996).
Gasser, R.B. et al. Analysis of sequence homogenisation in rDNA arrays of Haemonchus contortus by DGGE. Electrophoresis 19, 2391–2395 (1998).
Hayashi, K. & Yandell, D.W. How sensitive is PCR-SSCP? Human Mut. 2, 338–346 (1993).
Sheffield, V.C., Beck, J.S., Kwitek, A.E., Sandstrom, D.W. & Stone, E.M. The sensitivity of single strand conformation polymorphism analysis for the detection of single base substitutions. Genomics 16, 325–332 (1993).
Teschauer, W., Mussack, T., Braun, A., Waldner, H. & Fink, E. Conditions for single strand conformation polymorphism (SSCP) analysis with broad applicability: a study on the effects of acrylamide, buffer and glycerol concentrations in SSCP analysis of exons of the p53 gene. Eur. J. Clin. Chem. Clin. Biochem. 34, 125–131 (1996).
Zhu, X.Q. & Gasser, R.B. SSCP-based mutation scanning approaches to fingerprint sequence variation in ribosomal DNA of ascaridoid nematodes. Electrophoresis 19, 1366–1373 (1998).
Doi, K. et al. High-throughput single nucleotide polymorphism typing by fluorescent single-strand conformation polymorphism analysis with capillary electrophoresis. Electrophoresis 25, 833–838 (2004).
Jespersgaard, C. et al. Optimization of capillary array electrophoresis single-strand conformation polymorphism analysis for routine molecular diagnostics. Electrophoresis 27, 3816–3822 (2006).
Tahira, T. et al. QSNPlite, a software system for quantitative analysis of SNPs based on capillary array SSCP analysis. Electrophoresis 27, 3869–3878 (2006).
Gasser, R.B. & Chilton, N.B. Applications of single-strand conformation polymorphism (SSCP) to taxonomy, diagnosis, population genetics and molecular evolution of parasitic nematodes. Vet. Parasitol. 101, 201–213 (2001).
Gasser, R.B., Zhu, X.Q., Hu, M., Jacobs, D.E. & Chilton, N.B. Molecular Genetic Characterisation of Members of the Genus Toxocara (Nematoda: Ascaridoidea) – Systematic, Population Genetic and Epidemiological Considerations: in Toxocara: The Enigmatic Parasite (eds. Holland, C. & Smith, H.) (CABI Press, Wallingford, UK, 2005).
Cafarchia, C. et al. Multilocus mutation scanning for the analysis of genetic variation within Malassezia (Basidiomycota: Malasseziales). Electrophoresis (in the press).
Sunnucks, P. et al. SSCP is not so difficult: the application and utility of single-stranded conformation polymorphism in evolutionary biology and molecular ecology. Mol. Ecol. 9, 1699–1710 (2000).
King, S., McCord, B.R. & Riefler, R.G. Capillary electrophoresis single-strand conformation polymorphism analysis for monitoring soil bacteria. J. Microbiol. Methods 60, 83–92 (2005).
Garrick, R.C. & Sunnucks, P. Development and application of three-tiered nuclear genetic markers for basal Hexapods using single-stranded conformation polymorphism coupled with targeted DNA sequencing. BMC Genet. 7, 11 (2006).
Gasser, R.B. et al. Genotyping Cryptosporidium parvum by single-strand conformation polymorphism analysis of ribosomal and heat shock gene regions. Electrophoresis 22, 433–437 (2001).
Gasser, R.B., Abs EL-Osta, Y.G. & Chalmers, R.M. Electrophoretic analysis of genetic variability within Cryptosporidium parvum from imported and autochtonous cases of cryptosporidiosis in the United Kingdom. Appl. Environ. Microbiol. 69, 2719–2730 (2003).
Chalmers, R.M. et al. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis. Int. J. Parasitol. 35, 397–410 (2005).
Gasser, R.B., Hu, M., Abs EL-Osta, Y.G., Zarlenga, D.S. & Pozio, E. Non-isotopic single-strand conformation polymorphism analysis of sequence variability in ribosomal DNA expansion segments within the genus Trichinella (Nematoda: Adenophorea). Electrophoresis 25, 3357–3364 (2004).
Innis, M.A. & Gelfand, D.H. Optimization of PCRs. in PCR Protocols – A Guide to Methods and Applications (eds. Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J.) 3–12 (Academic Press, New York, 1990).
Acknowledgements
We would like to thank colleagues who have contributed to research in our laboratory. Our research has been supported largely through grants from the Australian Research Council, Genetic Technologies Limited, the Australian Academy of Science, Meat and Livestock Australia and, more recently, Elchrom Scientific AG. Approval has been granted by Wiley Press to reproduce partial images (presented in Fig. 2) from previous publications23,30.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Gasser, R., Hu, M., Chilton, N. et al. Single-strand conformation polymorphism (SSCP) for the analysis of genetic variation. Nat Protoc 1, 3121–3128 (2006). https://doi.org/10.1038/nprot.2006.485
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.485
This article is cited by
-
Genetic Characterization of Myf5 and POU1F1 Genes in Different Egyptian Local Rabbit Breeds and Their Association with Growth Traits
Biochemical Genetics (2023)
-
Equus roundworms (Parascaris univalens) are undergoing rapid divergence while genes involved in metabolic as well as anthelminic resistance are under positive selection
BMC Genomics (2022)
-
Association of PRLR, IGF1, and LEP genes polymorphism with milk production and litter size in Egyptian Zaraibi goat
Tropical Animal Health and Production (2022)
-
Rapid and optimized protocol for efficient PCR-SSCP genotyping for wide ranges of species
Biologia (2021)
-
A novel DNAH5 variant in a Tunisian patient with primary ciliary dyskinesia
Journal of Genetics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.