Abstract
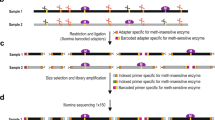
Restriction landmark genomic scanning (RLGS) is a method to detect large numbers of restriction landmarks in a single experiment. It is based on the concept that restriction enzyme sites can serve as landmarks throughout a genome. RLGS uses direct end-labeling of the genomic DNA digested with a rare-cutting restriction enzyme and high-resolution two-dimensional electrophoresis. Compared with the conventional gene-detection technologies, such as Southern blot analysis and PCR, RLGS has the following advantages even though it needs specially designed instruments: high-efficiency scanning capacity, scanning extensibility by using alternate restriction enzyme combinations, applicability to any organism, a spot intensity that reflects the copy number of restriction landmarks, and the ability, by using a methylation-sensitive enzyme, to screen the methylated state of genomic DNA. The RLGS protocol can be accomplished in 5 days to 2 weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hatada, I., Hayashizaki, Y., Hirotsune, S., Komatsubara, H. & Mukai, T. A genomic scanning method for higher organisms using restriction sites as landmarks. Proc. Natl. Acad. Sci. USA 88, 9523–9527 (1991).
Hayashizaki, Y. et al. Restriction landmark genomic scanning method and its various applications. Electrophoresis 14, 251–258 (1993).
Okazaki, Y. et al. A genetic linkage map of the mouse using an expanded production system of restriction landmark genomic scanning (RLGS Ver.1.8). Biochem. Biophys. Res. Commun. 205, 1922–1929 (1994).
Hayashizaki, Y., Hirotsune, S., Okazaki, Y., Muramatsu, M. & Ashikawa, J. Restriction landmark genomic scanning method. In Molecular Biology and Biotechnology: a comprehensive desk reference. (ed. Meyers, R.) 813–817 (VCH, Weinheim, 1995).
Hayashizaki, Y. & Watanabe, S., eds. Restriction landmark genomic scanning (RLGS). (Springer, Berlin, 1997).
Kawai, J., Hayashizaki, Y., Okazaki, Y., Suzuki, H. & Watanabe, S. Restriction landmark genomic and cDNA scanning. In Encyclopedia of Analytical Chemistry: Instrumentation and Applications. (ed. Meyers, R.) 5196–5222, (John Wiley & Sons Ltd, Chichester, 2000).
Southern, E.M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98, 503–517 (1975).
Saiki, R.K. et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230, 1350–1354 (1985).
Shi, H. et al. Oligonucleotide-based microarray for DNA methylation analysis: principles and applications. J. Cell. Biochem. 88, 138–143 (2003).
Asakawa, J. et al. Quantitative and qualitative genetic variation in two-dimensional DNA gels of human lymphocytoid cell lines. Electrophoresis 16, 241–252 (1995).
Kawai, J. et al. Methylation profiles of genomic DNA of mouse developmental brain detected by restriction landmark genomic scanning (RLGS) method. Nucleic Acids Res. 21, 5604–5608 (1993).
Dahl, C. & Guldberg, P. DNA methylation analysis techniques. Biogerontology 4, 233–250 (2003).
Akama, T.O. et al. Restriction landmark genomic scanning (RLGS-M)-based genome-wide scanning of mouse liver tumors for alterations in DNA methylation status. Cancer Res. 57, 3294–3299 (1997).
Watanabe, S. et al. Accessibility to tissue-specific genes from methylation profiles of mouse brain genomic DNA. Electrophoresis 16, 218–226 (1995).
Dai, Z. et al. An AscI boundary library for the studies of genetic and epigenetic alterations in CpG islands. Genome Res. 12, 1591–1598 (2002).
Hayashizaki, Y. et al. A genetic linkage map of the mouse using restriction landmark genomic scanning (RLGS). Genetics 138, 1207–1238 (1994).
Hirotsune, S. et al. Spot mapping on the standard profile of restriction landmark genomic scanning (RLGS) of sorted chromosome 20 using methylation-insensitive enzyme. Genomics 24, 593–596 (1994).
Okuizumi, H. et al. Genetic mapping of restriction landmark genomic scanning loci in the mouse. Electrophoresis 16, 233–240 (1995).
Hirotsune, S. et al. Construction of high-resolution physical maps from yeast artificial chromosomes using restriction landmark genomic scanning (RLGS). Genomics 37, 87–95 (1996).
Okazaki, Y. et al. A genetic linkage map of the Syrian hamster and localization of cardiomyopathy locus on chromosome 9qa2.1-b1 using RLGS spot-mapping. Nat. Genet. 13, 87–90 (1996).
Hayashizaki, Y. et al. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat. Genet. 6, 33–40 (1994).
Shibata, H. et al. The use of restriction landmark genomic scanning to scan the mouse genome for endogenous loci with imprinted patterns of methylation. Electrophoresis 16, 210–217 (1995).
Plass, C. et al. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat. Genet. 14, 106–109 (1996).
Costello, J.F. & Plass, C. Methylation matters. J. Med. Genet. 38, 285–303 (2001).
Rush, L.J. & Plass, C. Restriction landmark genomic scanning for DNA methylation in cancer: past, present, and future applications. Anal. Biochem. 307, 191–201 (2002).
Laird, P. W. & Jaenisch, R. DNA methylation and cancer. Hum. Mol. Genet. 3, 1487–1495 (1994).
Counts, J.L. & Goodman, J.I. Alterations in DNA methylation may play a variety of roles in carcinogenesis. Cell 83, 13–15 (1995).
Vachtenheim, J., Horakova, I. & Novotna, H. Hypomethylation of CCGG sites in the 3′ region of H-ras protooncogene is frequent and is associated with H-ras allele loss in non-small cell lung cancer. Cancer Res. 54, 1145–1148 (1994).
Ohtani-Fujita, N. et al. CpG methylation inactivates the promoter activity of the human retinoblastoma tumor-suppressor gene. Oncogene 8, 1063–1067 (1993).
Ottaviano, Y.L. et al. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 54, 2552–2555 (1994).
Asaga, S. et al. Identification of a new breast cancer-related gene by restriction landmark genomic scanning. Anticancer Res. 26, 35–42 (2006).
Costello, J.F. et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 24, 132–138 (2000).
Okazaki, Y. et al. Direct detection and isolation of restriction landmark genomic scanning (RLGS) spot DNA markers tightly linked to a specific trait by using the RLGS spot-bombing method. Proc. Natl. Acad. Sci. USA 92, 5610–5614 (1995).
Hirotsune, S. et al. Molecular cloning of polymorphic markers on RLGS gel using the spot target cloning method. Biochem. Biophys. Res. Commun. 194, 1406–1412 (1993).
Ohsumi, T. et al. A spot cloning method for restriction landmark genomic scanning. Electrophoresis 16, 203–209 (1995).
Hayashizaki, Y. et al. A new method for constructing NotI linking and boundary libraries using a restriction trapper. Genomics 14, 733–739 (1992).
Suzuki, H., Kawai, J., Taga, C., Ozawa, N. & Watanabe, S. A PCR-mediated method for cloning spot DNA on restriction landmark genomic scanning (RLGS) gel. DNA Res. 1, 245–250 (1994).
Plass, C. et al. An arrayed human Not I-EcoRV boundary library as a tool for RLGS spot analysis. DNA Res. 4, 253–255 (1997).
Smiraglia, D.J. et al. A new tool for the rapid cloning of amplified and hypermethylated human DNA sequences from restriction landmark genome scanning gels. Genomics 58, 254–262 (1999).
Akiyoshi, S. et al. A genetic linkage map of the MSM Japanese wild mouse strain with restriction landmark genomic scanning (RLGS). Mamm. Genome 11, 356–359 (2000).
Rouillard, J.M. et al. Virtual genome scan: a tool for restriction landmark-based scanning of the human genome. Genome Res. 11, 1453–1459 (2001).
Zardo, G. et al. Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nat. Genet. 32, 453–458 (2002).
Matsuyama, T. et al. Global methylation screening in the Arabidopsis thaliana and Mus musculus genome: applications of virtual image restriction landmark genomic scanning (Vi-RLGS). Nucleic Acids Res. 31, 4490–4496 (2003).
Takamiya, T. et al. Restriction landmark genome scanning method using isoschizomers (MspI/HpaII) for DNA methylation analysis. Electrophoresis 27, 2846–2856 (2006).
Acknowledgements
We thank J. Kawai for valuable comments. This work was supported by a Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ando, Y., Hayashizaki, Y. Restriction landmark genomic scanning. Nat Protoc 1, 2774–2783 (2006). https://doi.org/10.1038/nprot.2006.350
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.350
This article is cited by
-
An Overview of Epigenetic Assays
Molecular Biotechnology (2008)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.