Abstract
Converging evidence suggests that major depressive disorder (MDD) affects multiple large-scale brain networks. Analyses of the correlation or covariance of regional brain structure and function applied to structural and functional MRI data may provide insights into systems-level organization and structure-to-function correlations in the brain in MDD. This study applied tensor-based morphometry and intrinsic connectivity distribution to identify regions of altered volume and intrinsic functional connectivity in data from unmedicated individuals with MDD (n=17) and healthy comparison participants (HC, n=20). These regions were then used as seeds for exploratory anatomical covariance and connectivity analyses. Reduction in volume in the anterior cingulate cortex (ACC) and lower structural covariance between the ACC and the cerebellum were observed in the MDD group. Additionally, individuals with MDD had significantly lower whole-brain intrinsic functional connectivity in the medial prefrontal cortex (mPFC). This mPFC region showed altered connectivity to the ventral lateral PFC (vlPFC) and local circuitry in MDD. Global connectivity in the ACC was negatively correlated with reported depressive symptomatology. The mPFC–vlPFC connectivity was positively correlated with depressive symptoms. Finally, we observed increased structure-to-function correlation in the PFC/ACC in the MDD group. Although across all analysis methods and modalities alterations in the PFC/ACC were a common finding, each modality and method detected alterations in subregions belonging to distinct large-scale brain networks. These exploratory results support the hypothesis that MDD is a systems level disorder affecting multiple brain networks located in the PFC and provide new insights into the pathophysiology of this disorder.
Similar content being viewed by others
Introduction
Major depressive disorder (MDD) is a highly heterogeneous and potentially disabling disorder that affects multiple large-scale brain networks (Drevets, 2001; Nestler et al, 2002). A growing array of neuroimaging technical advances now makes possible the characterization of the structural and functional integrity of these large-scale networks in vivo in patients. For example, one can measure the covariance of brain structure on magnetic resonance imaging (MRI) to characterize putative anatomical covariance networks (Alexander-Bloch et al, 2013; Evans, 2013). Functional connectivity is defined in the correlation or covariance in regional brain activity over time (Craddock et al, 2013) as measured using functional MRI (fMRI). This approach has been applied to the characterization of human functional networks (Finn et al, 2015; Yeo et al, 2011). Within individuals, networks defined by anatomical covariance and intrinsic functional connectivity are networks similar to, but distinct from, each other (Clos et al, 2014).
Intrinsic functional connectivity studies in MDD have highlighted abnormalities in several large-scale functional networks including the default mode, frontoparietal, and salience networks (Drysdale et al, 2017; Kaiser et al, 2015; Mulders et al, 2015). The majority of these studies have focused on seed-based intrinsic functional connectivity, which requires a priori knowledge to select regions or networks of interest. The need for a priori knowledge limits the ability to identify novel disturbances. In contrast, data-driven, voxel-based methods, which examine the intrinsic functional connectivity of each voxel to every other voxel in the brain, are not constrained by this problem. Using this approach, two recent studies reported lower global connectivity in the medial prefrontal cortex (mPFC; Murrough et al, 2016; Wang et al, 2014).
MDD is associated with lower volume of brain regions including prefrontal cortex (PFC) and anterior cingulate cortex (ACC) (Hajek et al, 2008; Koolschijn et al, 2009). However, anatomical covariance studies are just emerging. For example, PFC thickness correlates with cortical thickness in other frontal association cortex regions (Qiu et al, 2014) and subcortical volumes (Zhao et al, 2017) in MDD. However, the direction of these alterations (whether thickening or thinning) differs between these studies. Similarly, studies employing whole-brain graph theory-based approaches differ on whether MDD is associated with increased (Ajilore et al, 2014; Mak et al, 2016), decreased (Wang et al, 2016), or no differences (Lim et al, 2013) in several graph theoretical measures. Further research is therefore needed to clarify the alterations of anatomical covariance in MDD and its relationship to symptoms in order to develop more efficacious interventions. The relationship between anatomical covariance and intrinsic functional connectivity in MDD is also unclear.
Here we report the application of tensor-based morphometry (TBM; Hua et al, 2008) and intrinsic connectivity distribution (ICD; Scheinost et al, 2012) to identify regions of altered volume and intrinsic functional connectivity in MDD as hypothesis-generating analyses. Regions identified as being altered were then used as seeds for exploratory anatomical covariance and seed connectivity analyses. Finally, we examined the correlation between these structural and functional alterations to detect potential shared variance between the two modalities. Based on existing literature (Hajek et al, 2008; Koolschijn et al, 2009; Murrough et al, 2016; Wang et al, 2014), we hypothesized that individuals with MDD would show lower volume and intrinsic functional connectivity in the mPFC and ACC, and that this might be related to the severity of depression. We had no a priori hypotheses for the follow-up seed analyses.
Materials and methods
All study procedures were approved by the Yale University Institutional Review Board and the Yale MRRC Protocol Review Committee. All participants provided written, informed consent.
Participants
Inclusion criteria were assessed by a physical examination, routine blood tests, and medical and psychiatric histories. A drug screen (urine), ECG, and a pregnancy test (for female participants) were performed as part of the subject screening. The Montgomery–Åsberg Depression Rating Scale (MADRS; Montgomery and Asberg, 1979) and the Hamilton Depression Rating Scale (HAMD-17; Hamilton, 1967) were used to assess participants’ mood both during intake and on MRI scan day. Inclusion criteria were: (1) aged between 18 and 60 years old, (2) English speaking, and (3) no lifetime drug abuse or dependence disorder (excluding nicotine use). Additional inclusion criteria for healthy comparison (HC) participants were: (1) no current, or history of, any DSM-IV diagnosis as assessed by the Structured Clinical Interview for DSM (SCID) besides nicotine dependence, (2) no first-degree relative with a history of psychotic, mood, or anxiety disorder, and (3) no regular medication use within the past 2 months, and no history of psychiatric medication use. Inclusion criteria for depressed participants were: (1) currently in a depressed mood state as defined by a MADRS score ⩾10 and DSM-IV SCID, (2) primary diagnosis of MDD as assessed by DSM-IV SCID, (3) no other current DSM-IV diagnosis, besides anxiety disorder and nicotine dependence, (4) no use of psychiatric medication in the past month preceding the study, and (5) no significant suicidal ideation. The DSM-IV SCID (APA, 2000) and rating scales were administered by trained researchers with experience in assessments of individuals with mood disorders. Diagnosis was then confirmed by a licensed clinical psychologist.
In total, 17 unmedicated MDD and 20 HC participants were scanned. The two groups did not significantly differ in terms of age, sex, or handedness. Individuals with MDD had significantly less education compared to HC participants. Subject characteristics are presented in Table 1.
Imaging Parameters
Participants were scanned on a Siemens 3T Tim Trio scanne. After a first localizing scan, a high-resolution 3D volume was collected using a magnetization prepared rapid gradient echo sequence (176 contiguous sagittal slices, slice thickness=1 mm, matrix =192 × 192, FoV=256 mm, repetition time (TR)=2530 ms, echo time (TE)=2.77 ms, flip angle=7°). A 2D T1-weighted image with the same slice prescription as the functional images was also collected for purposes of registration. Functional runs included 340 whole-brain volumes acquired using a multiband echo-planar imaging sequence with the following parameters: TR=1 s, TE=30 ms, flip angle=62°, matrix=84 × 84, in-plane resolution=2.5 mm2, 51 axial-oblique slices parallel to the ac–pc line, slice thickness=2.5, multiband=3, acceleration factor=2.
Tensor-Based Morphometry
First, images were skull stripped using FSL (https://fsl.fmrib.ox.ac.uk/fsl/) and any remaining non-brain tissue was manually removed. All further analyses were performed using BioImage Suite (Joshi et al, 2011) unless otherwise specified. Images were aligned to MNI space using a 12-parameter affine registration by maximizing the normalized mutual information between individual scans and the MNI template brain. These aligned images were averaged together to form the initial template for non-linear registration.
For TBM analysis, images were non-linearly registered to an evolving group average template in an iterative fashion using a previously validated algorithm (Scheinost et al, 2017). This algorithm iterates between estimating a local transformation to align individual brains to a group average template and creating a new group average template based on the previous transformations. The local transformation was modeled using a free-form deformation (FFD) parameterized by cubic B-splines. This transformation deforms an object by manipulating an underlying mesh of control points. The deformation for voxels in between control points was interpolated using B-splines to form a continuous deformation field. Positions of control points were optimized using conjugate gradient descent to maximize the normalized mutual information between the template and individual brains. After each iteration, the quality of the local transformation was improved by increasing the number of control points and decreasing the spacing between control points to capture a more precise alignment. A total of four iterations were performed with decreasing control point spacings of 15, 10, 5, and 2.5 mm. To help prevent local minimums during optimization, a multi-resolution approach was used and three resolution levels were used at each iteration.
The determinant of the Jacobian of the deformation field was used to quantify local volume differences between the registered images and the template (Hua et al, 2008). This metric provided an estimate of voxel-wise volume changes for all transformed images with respect to the group averaged template and was used for further analysis.
Exploratory Seed-Based Anatomical Covariance
Based on regions of significant volume differences, seed regions were chosen for anatomical covariance analysis as previously described (Scheinost et al, 2017). The volumes of these seed regions, calculated as the average determinant of the Jacobian in the seed, were extracted and used as covariates in linear models (described below).
Intrinsic Functional Connectivity Preprocessing
The first 10 volumes of each functional run were discarded to allow for the magnetization to reach a steady state. Motion correction was performed using SPM8 (http://www.fil.ion.ucl.ac.uk/spm/). Images were warped into common space using the non-linear transformation described above using cubic interpolation and were iteratively smoothed until the smoothness of any image had a full-width at half-maximum of ~5 mm (Scheinost et al, 2014) using AFNI’s 3dBlurToFWHM (http://afni.nimh.nih.gov/afni/). This iterative smoothing reduces motion related confounds. All further analyses were performed using BioImage Suite. Several covariates of no interest were regressed from the data including linear and quadratic drifts, mean cerebral-spinal-fluid (CSF) signal, mean white-matter signal, and mean gray matter signal. For additional control of possible motion related confounds, a 24-parameter motion model (including six rigid-body motion parameters, six temporal derivatives, and these terms squared) was regressed from the data. The data were temporally smoothed with a Gaussian filter (approximate cutoff frequency=0.12 Hz). A canonical gray matter mask defined in common space was applied to the data, so only voxels in the gray matter were used in further calculations. Finally, for each participant, all preprocessed resting-state runs were variance normalized and concatenated.
Intrinsic Connectivity Distribution
After preprocessing, intrinsic functional connectivity of each voxel as measured by the ICD was calculated for each subject as described previously (Scheinost et al, 2012). Similar to other data-driven, voxel-based measures (Murrough et al, 2016; Wang et al, 2014), ICD involves correlating the time series for any voxel with every other time series in the brain or brain hemisphere. A summary statistic based on the network theory measure degree is then calculated. ICD avoids the need for choosing an arbitrary connectivity threshold by modeling the entire distribution of correlation thresholds using a Weibull distribution:  , where r is a correlation between the two time series, α is the variance parameter, and β is the shape parameter. This parameterization is akin to modeling the change in network theory metric degree, as the threshold used to calculate degree is increased, with a stretched exponential:
, where r is a correlation between the two time series, α is the variance parameter, and β is the shape parameter. This parameterization is akin to modeling the change in network theory metric degree, as the threshold used to calculate degree is increased, with a stretched exponential:  , where τ is the correlation threshold, and α and β are the parameters as above.
, where τ is the correlation threshold, and α and β are the parameters as above.
Specifically, the time series for any gray matter voxel was correlated with every other voxel in the gray matter. A histogram of these correlations was constructed to estimate the distribution of connections to the current voxel. The survival function was estimated as 1−F(x), where F(x) is the cumulative sum of the histogram, and was fitted with a stretched exponential with unknown variance. As variance controls the spread of the distribution of connections, a larger variance indicates a greater number of high correlation connections. Finally, this process is repeated for all voxels in the gray matter, resulting in a whole-brain parametric image summarizing the connectivity of each tissue element.
Exploratory Follow-Up Seed Connectivity
Follow-up seed analysis was performed to explore (post hoc) the nodes identified by ICD analysis to determine the specific connections that were most responsible for changes in connectivity. The time series of the seed region in a given participant was then computed as the average time series across all voxels in the seed region. This time series was correlated with the time series for every other voxel in the gray matter to create a map of r-values, reflecting seed-to-whole-brain connectivity. These r-values were transformed to z-values using Fisher’s transform yielding one map for each participant representing the strength of correlation to the seed region.
Motion Analysis
As group differences in motion have been shown to confound connectivity studies, we calculated the average frame-to-frame displacement for each participant’s data. In line with current reports, participants with an average frame-to-frame displacement >0.10 for any run were removed from the analysis (2 MDD and 1 HC). There were no significant differences for motion between the MDD and HC groups (MDD: 0.06±0.02, HC: 0.05±0.03, p=0.16). Finally, we regressed a 24-parameter motion model and used an iterative smoothing algorithm to minimize any motion confounds (Satterthwaite et al, 2013; Scheinost et al, 2014).
Group Analysis
TBM and seed anatomical covariance data were analyzed with voxel-wise general linear modeling. Age at scan and sex were included as covariates. For models of anatomical covariance, seed volumes and a group-by-volume interaction were added as covariates of interest. Regions showing significant group-by-volume interaction are regions where the correlation between the volume of that region and the seed region is different between groups. Voxel-wise t-tests were used to compare the connectivity data between the study groups. Comparisons of the TBM and ICD data were considered primary analyses, while comparisons of the seed anatomical covariance and connectivity data were considered exploratory. Imaging results are shown at a cluster-level threshold of p<0.05 family-wise error (FWE) correction as determined by AFNI’s 3dClustSim program (version 16.0.09) using a cluster-forming threshold of p=0.001, 10 000 iterations, a gray matter mask, and a smoothness estimated from the residuals using 3dFWHMx with the –ACF option. This stringent cluster-forming threshold was used to avoid the problems of false positives associated with less stringent cluster-forming thresholds and is based on current best practices (Eklund et al, 2016; Nichols et al, 2017).
To investigate the structure-to-function correlation between TBM and ICD values, we used a high-resolution functional atlas (Finn et al, 2015) to divide the PFC, including the ACC, into 52 functional nodes as shown in Supplementary Figure S1. For each participant, the averaged TBM and ICD values in these nodes were computed, the average TBM and ICD values were correlated, and the resulting correlation was converted to a z-value via the Fisher transform to ensure normality. This approach resulted in a single structure-to-function correlation between TBM and ICD in the PFC for each participant. The structure-to-function correlations were compared across study groups using a two-sample t-test. Additionally, we repeated this analysis using nodes located outside the prefrontal cortex (n=216) and all nodes, covering the whole brain.
Results
Group Differences in Gray Matter Volume and Anatomical Covariance
Whole-brain analyses of gray matter volume are presented in Figure 1a. For individuals with MDD compared to HC participants, TBM analysis revealed significantly (p<0.05, corrected, degrees of freedom (df)=32) higher volume in the right middle and inferior temporal gyri (BA 20) and lower volume in the dorsal ACC (dACC). No significant correlations between the TBM data and either MADRS or HAMD-17 were observed.
Individuals with major depressive disorder (MDD) exhibited altered gray matter volume and covariance. (a) Individuals with MDD exhibited increased volume in the right middle and inferior temporal gyri and reduced volume in the dorsal anterior cingulate (dACC) compared to healthy comparison (HC) participants. When these clusters were used as seeds for anatomical covariance analysis, individuals with MDD compared to HC participants exhibited significantly (p<0.05, corrected) (b) increased covariance between the middle/inferior temporal gyri and the orbital frontal cortex (OFC), and (c) decreased covariance between the dACC and the vermis in the anterior lobe of the cerebellum.
These two clusters were then used as seeds for anatomical covariance analysis to interrogate if volume differences were associated with additional network level differences. When using the right middle/inferior temporal gyri cluster as a seed, MDD compared to HC participants exhibited significantly (p<0.05, corrected, df=30) higher covariance with the orbital frontal cortex (OFC). There was a significant positive covariance in volume between the two regions in the MDD (p<0.001, df=15) but not the HC group (p=0.67, df=18). When using the dACC cluster, there was a significantly (p<0.05, corrected, df=30) lower covariance with the vermis in the anterior lobe of the cerebellum in the MDD but not HC group. In the MDD group, there was a trend toward significant negative covariance in volume between the two regions (p=0.08, df=15) while in the HC group there was a significant positive covariance (p<0.001, df=18). Scatterplots of these covariance is shown in Supplementary Figure S2.
Group Differences in ICD Connectivity and Associations with Symptoms
Single group ICD maps and whole-brain comparison of ICD connectivity between the MDD and HC participants are shown in Figure 2a–c. ICD connectivity in the vmPFC was significantly (p<0.05 corrected, df=32) lower in the MDD as compared to the HC group. No regions of higher ICD connectivity for MDD compared to HC groups were detected.
Individuals with major depressive disorder (MDD) exhibited lower intrinsic functional connectivity in the medial prefrontal cortex (mPFC). Single group ICD maps for (a) HC participants and (b) individuals with MDD show greatest connectivity in midline ‘hub’ regions with visually reduced ICD connectivity in the vmPFC for the MDD patients. (c) For MDD compared to HC, the vmPFC exhibited significantly (p<0.05 corrected) reduced ICD connectivity. (d) For individuals with MDD, ICD connectivity in the ventral anterior cingulate cortex (vACC) was inversely correlated with Hamilton Depression Rating Scale (1st 17). Images shown at p<0.05, corrected for multiple comparisons.
For the MDD group, ICD connectivity in the ventral anterior cingulate cortex (vACC) was inversely correlated (p<0.05 corrected, df=12) with depressive symptoms on the HAMD-17 (Figure 2d) in a consistent direction with the between-group differences in Figure 2c. A scatterplot of this correlation is shown in Supplementary Figure S3A.
Group Difference in Seed Connectivity and Associations with Symptoms
Follow-up seed connectivity from the vmPFC regions detected from the ICD analysis (Figure 2c) is shown in Figure 3. For MDD compared to HC, seed connectivity using the vmPFC seed revealed significantly (p<0.05 corrected, df=32) higher connectivity to the ventral lateral PFC (vlPFC) and lower connectivity to the anterior PFC (aPFC).
Individuals with MDD showed altered connectivity for the vmPFC region detected from the ICD analysis (Figure 2c) to other regions of the PFC. (a) For HC participants, the vmPFC was significantly (p<0.05 corrected) correlated with regions of the default mode network (DMN) and negatively correlated with regions of the frontoparietal network (FPN), visual network, and the salience network (SAN). (b) For individuals with MDD, the vmPFC was significantly (p<0.05 corrected) connected to similar networks albeit at reduced strength. (c) For MDD compared to HC, seed connectivity revealed significantly increased (or significantly less negative) connectivity between the vmPFC and ventral lateral PFC and significantly reduced connectivity between the vmPFC and anterior prefrontal cortex.
For both clusters of significant between-group differences, connectivity values were extracted and associated with symptom severity. mPFC–vlPFC connectivity was positively correlated with depressive symptoms on both the MADRS (r=0.65, p=0.01, df=12) and the HAMD-17 (r=0.56, p=0.04, df=12). A scatterplot of these correlations are shown in Supplementary Figure S3B and C. No significant associations with symptom severity were observed for vmPFC–aPFC connectivity.
Network-Level Summary of Group Differences
As we hypothesized that our multimodal methods would detect group differences in several large-scale brain networks, we localized our results into seven networks using the cortical parcellation (Yeo et al, 2011) and the cerebellar parcellation (Buckner et al, 2011). Table 2 shows a summary of our results. Briefly, the TBM and anatomical covariance results were localized to the salience and the default mode networks. The ICD results were localized to the default mode network. The seed connectivity results were localized to the default mode and frontoparietal networks.
Group Difference in Structure-to-Function Correlation Between Prefrontal Volume and Prefrontal Connectivity
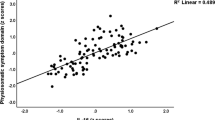
Given that both structural and functional differences were localized largely to the PFC, we hypothesized that the structure-to-function correlation in the PFC would be altered in individuals with MDD compared to HC. We observed a significantly greater correlation between connectivity and volume in the PFC for MDD compared to HC (MDD vs HC: t=2.25, p=0.03, df=30), which was due to a significant correlation between connectivity and volume in MDD (z(r)=0.39, p<0.001, df=14) but a trend toward significance in HC (z(r)=0.14, p=0.06, df=18). This increased structure-to-function correlation was localized to the PFC as no differences in correlations were detected across the whole brain (MDD: z(r)=0.17; HC: z(r)=0.12; MDD vs HC: t=1.0, p=0.31, df=30) or nodes not in the PFC (MDD: z(r)=0.15; HC: z(r)=0.11; MDD vs HC: t=0.9, p=0.36, df=30).
Discussion
In this exploratory, multimodal imaging study, we examined structural and functional network alterations in MDD. Structural and anatomical covariance differences were detected in the salience (ACC and vermis of the cerebellum) and default mode networks (OFC and temporal gyri), while intrinsic functional connectivity differences were detected in the frontoparietal (vlPFC) and default mode networks (mPFC). Specifically, we detected significantly lower volume in the anterior cingulate cortex (ACC) and lower anatomical covariance between the ACC and the cerebellum in the MDD group. Furthermore, there was significantly lower whole-brain intrinsic functional connectivity in the medial prefrontal cortex (mPFC). Using follow-up seed connectivity, this mPFC region showed altered connectivity to the ventral lateral PFC (vlPFC) and local circuitry in MDD. The dysregulation in intrinsic functional connectivity findings was shown to be associated with severity of depression. Finally, we observed higher structure-to-function correlation in the PFC/ACC in the MDD group. Together, these results point to alterations in multiple brain networks centered around the PFC in individuals with MDD.
Alterations in the PFC/ACC were a common finding across all analysis methods (TBM, anatomical covariance, ICD, and seed-based connectivity). Yet, each method detected different large-scale networks, supporting the hypothesis that MDD is a systems level disorder affecting multiple intertwined brain networks (Drevets, 2001; Drysdale et al, 2017; Nestler et al, 2002). Multiple large-scale brain networks extend into the PFC/ACC (Finn et al, 2015; Yeo et al, 2011) with each of these networks supporting diverse, often antagonistic, roles in cognition (Fox et al, 2005; Riva-Posse et al, 2014). For example, major nodes from the default mode, frontoparietal, and salience networks were identified in our analyses and the interactions between these networks form the basis of several neurocognitive models of MDD (Drysdale et al, 2017; Kaiser et al, 2015; Zheng et al, 2015). That the observed changes in structure-to-function correlation were specific to the PFC in the MDD groups suggests that alterations within any of these networks or modalities are related. The relevance of the distinct PFC networks identified in the current analysis for the clinical presentation or treatment of MDD remains to be determined.
The TBM analysis revealed lower ACC volume and higher right middle/inferior temporal gyri volume. The ACC finding is in line with a recent study from the ENIGMA Major Depressive Disorder Working Group (Schmaal et al, 2017) and the mounting evidence indicating a central role for the ACC in the pathophysiology of MDD (Diener et al, 2012). When seeded for anatomical covariance analysis, the ACC exhibited differential covariance with vermis volume in the anterior lobe of the cerebellum in MDD vs controls. This part of the cerebellum is structurally and functionally connected to the ACC as part of the salience, or ventral attention, network (Buckner et al, 2011; Krienen and Buckner, 2009). Such volumetric differences may therefore be related to the network level alterations previously observed in MDD (Kaiser et al, 2015). Additionally, recent reviews have highlighted converging evidence of vermis alterations in MDD and their possible involvement with both motor and cognitive alterations (Konarski et al, 2005; Phillips et al, 2015). Volume differences in the temporal lobes are not generally reported in MDD, outside of a few studies (Caetano et al, 2004; Kumar et al, 1998). However, when seeded, the middle/inferior temporal gyri showed altered covariance to the OFC. Both the OFC and temporal gyri are part of secondary portions of the default mode network (Andrews-Hanna, 2012; Yeo et al, 2011) and may represent additional alteration of the default mode network beyond those in the main hubs of the network (ie, the posterior cingulate cortex and mPFC) that are reported here and elsewhere (Kaiser et al, 2015; Mulders et al, 2015; Murrough et al, 2016; Wang et al, 2014).
Our data-driven intrinsic functional connectivity analysis revealed lower connectivity in the mPFC to the rest of the brain in the MDD group. As the mPFC is a core node of the default mode network (Andrews-Hanna, 2012; Yeo et al, 2011), these connectivity results suggest an alteration of the default mode network in MDD. Altered default mode connectivity is commonly reported in MDD and, due to the default mode network’s role in self-referential processing (Sheline et al, 2009), is often related to negative self-referential thinking (Kaiser et al, 2015; Mulders et al, 2015; Sheline et al, 2009).
Our ICD results replicate the finding of the two most similar studies based on measures of network centrality (Murrough et al, 2016; Wang et al, 2014). This is notable as our MDD group was unmedicated and one of previous studies used a mix of medicated and unmedicated participants (Wang et al, 2014). Anti-depressants can affect intrinsic functional connectivity data (McCabe and Mishor, 2011), which could limit replication. While the previous studies detected larger clusters of decreased connectivity covering a majority of the mPFC and ACC, we detected a more focal cluster. This is most likely due to our use of more conservative cluster correction methods, which are based on current best practices (Nichols et al, 2017). Reducing our statistical thresholds to previous standards, we detect more widespread results including a cluster of reduced connectivity in the vACC that overlaps the cluster of significant correlation between ICD and depressive symptoms (Figure 2d). Differences between these studies based on network centrality and other studies, for example (Drysdale et al, 2017), is likely methodological. Our study is unique in that we performed follow-up seed analysis on the mPFC cluster and exploring structure-to-connectivity correlations.
When seeded for follow-up analysis, the vmPFC region displayed increased connectivity to the vlPFC, which was associated with more severe depressive symptoms. The vlPFC is involved in external monitoring and focused attention as part of the frontoparietal network, whereas the mPFC is involved in internal attention and self-awareness as part of the default mode network (Andrews-Hanna, 2012). These networks are traditionally anticorrelated (Fox et al, 2005), but their activity was not correlated in the MDD group. The lack of functional antagonism between these networks may reflect a reduction in cortical efficiency (ie, a deficit in the suppression of neural noise) or perhaps contamination of functions of one network by input from the other, ie, bias towards internal thoughts over engaging with the external world (Anticevic et al, 2012a, b; Kaiser et al, 2015). For example, self-focused rumination, a core feature of depression (Roberts et al, 1998), could be related to a reduced ability to efficiently separate the default mode and frontoparietal networks.
There are a number of important strengths to this work. We used multimodal MRI to investigate network level alterations in MDD. This study of MDD reports shared variance between volume and intrinsic functional connectivity in the PFC for individuals with MDD, but not HC participants. For functional imaging, we used high spatial and temporal resolution fMRI data in comparison to most prior studies of MDD. Furthermore, in contrast to the historical 5 min of data, all participants had ~12 min of resting-state data that has been shown to increase reliability and power (Noble et al, 2017). The individuals with MDD were unmedicated at the time of scanning (for at least 1 month), which is a notable strength as antidepressants have been shown to affect intrinsic functional connectivity data (McCabe and Mishor, 2011) and previous studies have used a mix of medicated and unmedicated participants (Wang et al, 2014).
Our study has several limitations. Our sample size was modest; yet, we were able to detect significant between-group differences using more stringent current best practices for multiple comparison correction (Nichols et al, 2017). Furthermore, while we show a correlation between volume and connectivity in the PFC in the MDD group, our correlation analysis cannot assign causality. As such, we cannot determine if the structural or functional changes preceded one or the other or whether these alterations reflect a vulnerability factor for, or a consequence of, MDD. Thus, longitudinal studies incorporating MRI across multiple time points would be needed to elucidate the orgins of these alterations in MDD. Finally, given that we performed several different analyses on the same cohort, our results should be interpreted as hypothesis-generating rather than confirmatory.
In conclusion, this novel study used multi-modal imaging and system-level methods to detect depression-related differences in multiple large-scale networks centered around the PFC/ACC. Structural and anatomical covariance differences were detected in the salience and default mode networks; while intrinsic functional connectivity differences were detected in the frontoparietal and default mode networks. Additionally, for MDD, volume and intrinsic functional connectivity in the PFC may not be independent. Our findings provide further evidence that MDD is a systems level disorder affecting multiple brain networks. Tentatively, these data suggest that further work to identify treatments specifically targeting multiple networks, and their interactions, for depressive symptoms is warranted.
Funding and disclosure
Support provided by K01MH092681 (IE), VA National Center for PTSD (IE; JHK), Nancy Taylor Foundation (IE), YCCI (IE). JHK acknowledges the following relevant financial interests. He is a co-sponsor of a patent for the intranasal administration of ketamine for the treatment of depression that was licensed by Janssen Pharmaceuticals, the maker of s-ketamine. He has a patent related to the use of riluzole to treat anxiety disorders that was licensed by Biohaven Medical Sciences. He has stock or stock options in Biohaven Medical Sciences, ARett Pharmaceuticals, Blackthorn Therapeutics, and Luc Therapeutics. He consults broadly to the pharmaceutical industry, but his annual income over the past year did not exceed $5000 for any organization. He receives over $5000 in income from the Society of Biological Psychiatry for editing the journal Biological Psychiatry. He has fiduciary responsibility for the International College of Neuropsychopharmacology as president of this organization. AA disclosed that he is a member of the Scientific Advisory Board of and a consultant for BlackThorn Therapeutics. CGA has served as a consultant and/or on the advisory boards of Genentech and Janssen. The remaining authors declare no conflict of interest.
References
Ajilore O, Lamar M, Leow A, Zhang A, Yang S, Kumar A (2014). Graph theory analysis of cortical-subcortical networks in late-life depression. Am J Geriatr Psychiatry 22: 195–206.
Alexander-Bloch A, Giedd JN, Bullmore E (2013). Imaging structural co-variance between human brain regions. Nat Rev Neurosci 14: 322–336.
Andrews-Hanna JR (2012). The brain’s default network and its adaptive role in internal mentation. Neuroscientist 18: 251–270.
Anticevic A, Cole MW, Murray JD, Corlett PR, Wang XJ, Krystal JH (2012a). The role of default network deactivation in cognition and disease. Trends Cogn Sci 16: 584–592.
Anticevic A, Gancsos M, Murray JD, Repovs G, Driesen NR, Ennis DJ et al (2012b). NMDA receptor function in large-scale anticorrelated neural systems with implications for cognition and schizophrenia. Proc Natl Acad Sci USA 109: 16720–16725.
American Psychiatric Association (2000). Diagnostic and Statistical Manual of Mental Disorders (DSM – IV)—text revision. American Psychiatric Association: Washington, DC.
Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BT (2011). The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol 106: 2322–2345.
Caetano SC, Hatch JP, Brambilla P, Sassi RB, Nicoletti M, Mallinger AG et al (2004). Anatomical MRI study of hippocampus and amygdala in patients with current and remitted major depression. Psychiatry Res 132: 141–147.
Clos M, Rottschy C, Laird AR, Fox PT, Eickhoff SB (2014). Comparison of structural covariance with functional connectivity approaches exemplified by an investigation of the left anterior insula. Neuroimage 99: 269–280.
Craddock RC, Jbabdi S, Yan CG, Vogelstein JT, Castellanos FX, Di Martino A et al (2013). Imaging human connectomes at the macroscale. Nat Methods 10: 524–539.
Diener C, Kuehner C, Brusniak W, Ubl B, Wessa M, Flor H (2012). A meta-analysis of neurofunctional imaging studies of emotion and cognition in major depression. Neuroimage 61: 677–685.
Drevets WC (2001). Neuroimaging and neuropathological studies of depression: implications for the cognitive-emotional features of mood disorders. Curr Opin Neurobiol 11: 240–249.
Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y et al (2017). Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med 23: 28–38.
Eklund A, Nichols TE, Knutsson H (2016). Cluster failure: Why fMRI inferences for spatial extent have inflated false-positive rates. Proc Natl Acad Sci USA 113: 7900–7905.
Evans AC (2013). Networks of anatomical covariance. Neuroimage 80: 489–504.
Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM et al (2015). Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci 18: 1664–1671.
Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME (2005). The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci USA 102: 9673–9678.
Hajek T, Kozeny J, Kopecek M, Alda M, Höschl C (2008). Reduced subgenual cingulate volumes in mood disorders: a meta-analysis. J Psychiatry Neurosci 33: 91–99.
Hamilton M (1967). Development of a rating scale for primary depressive illness. Br J Soc Clin Psychol 6: 278–296.
Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW et al (2008). Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage 43: 458–469.
Joshi A, Scheinost D, Okuda H, Belhachemi D, Murphy I, Staib LH et al (2011). Unified framework for development, deployment and robust testing of neuroimaging algorithms. Neuroinformatics 9: 69–84.
Kaiser RH, Andrews-Hanna JR, Wager TD, Pizzagalli DA (2015). Large-scale network dysfunction in major depressive disorder: a meta-analysis of resting-state functional connectivity. JAMA Psychiatry 72: 603–611.
Konarski JZ, McIntyre RS, Grupp LA, Kennedy SH (2005). Is the cerebellum relevant in the circuitry of neuropsychiatric disorders? J Psychiatry Neurosci 30: 178–186.
Koolschijn PC, van Haren NE, Lensvelt-Mulders GJ, Hulshoff Pol HE, Kahn RS (2009). Brain volume abnormalities in major depressive disorder: a meta-analysis of magnetic resonance imaging studies. Hum Brain Mapp 30: 3719–3735.
Krienen FM, Buckner RL (2009). Segregated fronto-cerebellar circuits revealed by intrinsic functional connectivity. Cereb Cortex 19: 2485–2497.
Kumar A, Jin Z, Bilker W, Udupa J, Gottlieb G (1998). Late-onset minor and major depression: early evidence for common neuroanatomical substrates detected by using MRI. Proc Natl Acad Sci USA 95: 7654–7658.
Lim HK, Jung WS, Aizenstein HJ (2013). Aberrant topographical organization in gray matter structural network in late life depression: a graph theoretical analysis. Int Psychogeriatr 25: 1929–1940.
Mak E, Colloby SJ, Thomas A, O'Brien JT (2016). The segregated connectome of late-life depression: a combined cortical thickness and structural covariance analysis. Neurobiol Aging 48: 212–221.
McCabe C, Mishor Z (2011). Antidepressant medications reduce subcortical-cortical resting-state functional connectivity in healthy volunteers. Neuroimage 57: 1317–1323.
Montgomery SA, Asberg M (1979). A new depression scale designed to be sensitive to change. Br J Psychiatry 134: 382–389.
Mulders PC, van Eijndhoven PF, Schene AH, Beckmann CF, Tendolkar I (2015). Resting-state functional connectivity in major depressive disorder: a review. Neurosci Biobehav Rev 56: 330–344.
Murrough JW, Abdallah CG, Anticevic A, Collins KA, Geha P, Averill LA et al (2016). Reduced global functional connectivity of the medial prefrontal cortex in major depressive disorder. Hum Brain Mapp 37: 3214–3223.
Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, Monteggia LM (2002). Neurobiology of depression. Neuron 34: 13–25.
Nichols TE, Das S, Eickhoff SB, Evans AC, Glatard T, Hanke M et al (2017). Best practices in data analysis and sharing in neuroimaging using MRI. Nat Neurosci 20: 299–303.
Noble S, Scheinost D, Finn ES, Shen X, Papademetris X, McEwen SC et al (2017). Multisite reliability of MR-based functional connectivity. Neuroimage 146: 959–970.
Phillips JR, Hewedi DH, Eissa AM, Moustafa AA (2015). The cerebellum and psychiatric disorders. Front Public Health 3: 66.
Qiu L, Lui S, Kuang W, Huang X, Li J, Zhang J et al (2014). Regional increases of cortical thickness in untreated, first-episode major depressive disorder. Transl Psychiatry 4: e378.
Riva-Posse P, Choi KS, Holtzheimer PE, McIntyre CC, Gross RE, Chaturvedi A et al (2014). Defining critical white matter pathways mediating successful subcallosal cingulate deep brain stimulation for treatment-resistant depression. Biol Psychiatry 76: 963–969.
Roberts JE, Gilboa E, Gotlib IH (1998). Ruminative response style and vulnerability to episodes of dysphoria: gender, neuroticism, and episode duration. Cogn Ther Res 22: 401–423.
Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME et al (2013). An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage 64: 240–256.
Scheinost D, Benjamin J, Lacadie CM, Vohr B, Schneider KC, Ment LR et al (2012). The intrinsic connectivity distribution: a novel contrast measure reflecting voxel level functional connectivity. NeuroImage 62: 1510–1519.
Scheinost D, Kwon SH, Lacadie C, Vohr BR, Schneider KC, Papademetris X et al (2017). Alterations in anatomical covariance in the prematurely born. Cereb Cortex 27: 534–543.
Scheinost D, Papademetris X, Constable RT (2014). The impact of image smoothness on intrinsic functional connectivity and head motion confounds. Neuroimage 95: 13–21.
Schmaal L, Hibar DP, Sämann PG, Hall GB, Baune BT, Jahanshad N et al (2017). Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol Psychiatry 22: 900–909.
Sheline YI, Barch DM, Price JL, Rundle MM, Vaishnavi SN, Snyder AZ et al (2009). The default mode network and self-referential processes in depression. Proc Natl Acad Sci USA 106: 1942–1947.
Wang L, Dai Z, Peng H, Tan L, Ding Y, He Z et al (2014). Overlapping and segregated resting-state functional connectivity in patients with major depressive disorder with and without childhood neglect. Hum Brain Mapp 35: 1154–1166.
Wang T, Wang K, Qu H, Zhou J, Li Q, Deng Z et al (2016). Disorganized cortical thickness covariance network in major depressive disorder implicated by aberrant hubs in large-scale networks. Sci Rep 6: 27964.
Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M et al (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106: 1125–1165.
Zhao K, Liu H, Yan R, Hua L, Chen Y, Shi J et al (2017). Altered patterns of association between cortical thickness and subcortical volume in patients with first episode major depressive disorder: a structural MRI study. Psychiatry Res 260: 16–22.
Zheng H, Xu L, Xie F, Guo X, Zhang J, Yao L et al (2015). The altered triple networks interaction in depression under resting state based on graph theory. Biomed Res Int 2015: 386326.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Scheinost, D., Holmes, S., DellaGioia, N. et al. Multimodal Investigation of Network Level Effects Using Intrinsic Functional Connectivity, Anatomical Covariance, and Structure-to-Function Correlations in Unmedicated Major Depressive Disorder. Neuropsychopharmacol. 43, 1119–1127 (2018). https://doi.org/10.1038/npp.2017.229
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2017.229
This article is cited by
-
Brain connectivity in major depressive disorder: a precision component of treatment modalities?
Translational Psychiatry (2023)
-
Imaging synaptic density in depression
Neuropsychopharmacology (2023)
-
Graph theory analysis of whole brain functional connectivity to assess disturbances associated with suicide attempts in bipolar disorder
Translational Psychiatry (2022)
-
Beyond noise to function: reframing the global brain activity and its dynamic topography
Communications Biology (2022)
-
Intrinsic connectivity of the prefrontal cortex and striato-limbic system respectively differentiate major depressive from generalized anxiety disorder
Neuropsychopharmacology (2021)