Abstract
Major depressive disorder (MDD) is a debilitating and widespread illness that exerts significant personal and socioeconomic consequences. Recent genetic and brain-imaging studies suggest that bicaudal C homolog 1 gene (BICC1), which codes for an RNA-binding protein, may be associated with depression. Here, we show that BICC1 mRNA is upregulated in the dorsolateral prefrontal cortex and dentate gyrus of human postmortem MDD patients. We also show that BICC1 is increased in the prefrontal cortex and hippocampus in the rat chronic unpredictable stress (CUS) model of depression. In addition, we show in vivo that a single acute antidepressant dose of ketamine leads to a rapid decrease of BICC1 mRNA, while in vitro, we show that this is likely due to neuronal activity-induced downregulation of BICC1. Finally, we show that BICC1 knockdown in the hippocampus protects rats from CUS-induced anhedonia. Taken together, these findings identify a role for increased levels of BICC1 in the pathophysiology of depressive behavior.
Similar content being viewed by others
INTRODUCTION
Major depressive disorder (MDD) is a recurring psychiatric illness that affects up to 17% of the American population and can be severely debilitating by affecting sleep, work, social relationships, and appetite (Kessler et al, 2005). Although medications to relieve depressive symptoms exist, there is a time lag of weeks to months, and some patients receive no benefit from these drugs and are considered ‘treatment-resistant’. Clinical and preclinical studies demonstrate that prolonged stress and MDD are associated with neuronal atrophy of cortical and limbic brain regions, including the prefrontal cortex (PFC) and hippocampus (Drevets et al, 1997; Izquierdo et al, 2006; Kang et al, 2012; Radley et al, 2004, 2006; Rajkowska et al, 1999; Shansky et al, 2009; Willner, 2005), but the exact molecular mechanisms underlying MDD have not yet been identified. However, an understanding of these mechanisms is critical to the development of new potential molecular targets for the pharmacological treatment of MDD.
Studies to elucidate the pathophysiology of depression have included analysis of the genetic basis of MDD, focusing on single nucleotide polymorphisms (SNPs) that are associated with susceptibility to mood disorders, and have identified a number of SNPs associated with depression. One recent genome-wide association study (GWAS) identified two SNPs in the bicaudal C homolog 1 (BICC1) gene showing suggestive significance for association in depressed patients compared to psychiatrically healthy controls (Lewis et al, 2010). In addition, a recent structural and functional imaging study corroborated this association with the finding that the minor T allele of the BICC1 SNP showed a protective role against MDD and the accompanying structural and functional changes in hippocampus and other limbic brain regions (Bermingham et al, 2012).
BICC1 was first identified in Drosophila as a gene required for anterior–posterior patterning of the oocyte (Mahone et al, 1995) and is expressed widely in peripheral tissues and in all brain regions (Lewis et al, 2010). BICC1 is an RNA-binding protein that plays a role in cytoskeletal organization and cell-to-cell communication (Chicoine et al, 2007; Mahone et al, 1995; Snee and Macdonald, 2009). Further, via its sterile alpha motif (SAM) domain, BICC1 can block Wnt signaling (Kraus et al, 2012), a pathway implicated in the pathophysiology of depression (Voleti et al, 2011), by uncoupling disheveled-2 signaling from the canonical Wnt pathway (Maisonneuve et al, 2009). BICC1 is well-studied in the context of development (Maisonneuve et al, 2009) and polycystic kidney disease (Guay-Woodford, 2003), but there have been few studies of BICC1 in the brain.
In the present study, we sought to understand whether BICC1 expression in the brain regulates depressive behavior. Here, we show that BICC1 mRNA is upregulated in the dorsolateral prefrontal cortex (dlPFC) and dentate gyrus (DG) of human postmortem MDD patients, and consistent with this, BICC1 mRNA is increased in PFC and hippocampus in the rat chronic unpredictable stress (CUS) model of depression. We also show that BICC1 is rapidly and robustly downregulated following a single acute antidepressant dose of ketamine in vivo; in vitro, we show that this may be due to a rapid, activity-regulated degradation of BICC1 mRNA. Finally, we show that small hairpin RNA (shRNA)-mediated BICC1 knockdown in the hippocampus protects rats from CUS-induced anhedonia. Taken together, these findings suggest that elevated levels of BICC1 contribute to the pathophysiology of depressive behavior.
MATERIALS AND METHODS
Human Subjects
Brain tissue was collected as previously described from MDD and psychiatrically healthy matched subjects (Duric et al, 2013; Kang et al, 2007; Sibille et al, 2011). Informed consent was obtained from the next of kin for all subjects, and all procedures were approved by the University of Pittsburgh’s Committee for the Oversight of Research Involving the Dead and Institutional Review Board for Biomedical Research, or by the Institutional Review Board of the University Hospitals of Cleveland. Briefly, the right hemisphere of the brain was blocked coronally, then immediately frozen and stored at −80°C. For dlPFC samples, 20-μm sections were cut serially then collected in tubes with Trizol reagent and purified by RNeasy columns (Qiagen, USA), and RNA integrity was measured using RIN. For hippocampal samples, 60 μm sections were cut serially, micropunched from the DG and CA1 pyramidal cell layers, and RNA was extracted with RNAqueous kit (Ambion, USA) and cleaned up with RNeasy MinElute kit (Qiagen). Based on availability, hippocampal samples were from the Cleveland cohort, whereas the PFC samples were from both the Cleveland and Pittsburgh cohorts. Further analysis revealed no significant difference in BICC1 regulation (as measured by fold change of MDD compared to control) between the two regionally different cohorts.
Animals
Male Sprague-Dawley rats with initial weights of 175–250 g were pair-housed and maintained in standard conditions with a 12-h light/dark cycle. Rats were handled before use, and food and water were provided ad libitum throughout the experiments except when noted. Animal use and procedures were in accordance with the National Institutes of Health guidelines and approved by the Yale University Animal Care and Use Committee.
Quantitative Real-Time PCR
RNA was purified using RNAqueous (Ambion). cDNA was synthesized using dNTP primers and reverse transcriptase (Genisphere, USA). Gene- and species-specific primers for BICC1 were designed using Primer 3 software, and the qPCR was run on the Eppendorf Realplex Mastercycler or the Applied Biosystems 7900HT using SYBR Green (Qiagen). Ct values of genes of interest were normalized to that of housekeeping genes (HMBS or GAPDH). For postmortem studies, fold change of each MDD patient was calculated relative to its paired control.
CUS Procedures
Rats were age- and weight-matched prior to commencement of CUS. They were subjected to a sequence of 11 stressors over the course of 21 days. These stressors were cold (4°C for 1 h), cage rotation (1 h), isolation overnight, food or water deprivation overnight, light on overnight, light off during day (3 h), odor overnight, stroboscope overnight, crowding overnight, or tilted cage overnight (45°). All stressors were randomly interspersed throughout the stress period. For the 1-day mild stress group, rats were subjected to only the final day of stressors used in the 21-day CUS protocol.
Immobilization Stress
Rats were restrained in tapered plastic film tubes (DecapiCones, Braintree Scientific, USA) for 2 h.
Drug Administration
Rats were given a single intraperitoneal (i.p.) injection of 10 mg/kg ketamine (Fort Dodge, USA) or saline. In other experiments, rats were given chronic i.p. injection of 5 mg/kg fluoxetine hydrochloride (Lilly, Indianapolis, IN) or saline for 21 days.
Sucrose Preference Test
Rats were habituated to 1% sucrose for 48 h, with the side of the bottle counterbalanced across animals and days. On testing day, rats were water-deprived for 6 h, then presented with pre-weighed, identical bottles of 1% sucrose and water. Again, the side of the bottle was counterbalanced across animals. One hour later, the bottles were removed and weighed to determine consumption of each fluid. Results were averaged across 2 days of testing.
Forced Swim Test
Rats were subjected to one 15-min session of swimming in water. Sessions were scored off-line by a blind observer. Immobility was defined as the least amount of movement possible to stay afloat.
Novelty Suppressed Feeding Test
Rats were food-deprived overnight, then placed in an open field in the dark with pellets of food in the center. Time elapsed until the first bite of food was recorded as latency to feed. Immediately after the novelty test, home-cage feeding was performed as a control measure to verify that motivation for food was present.
Elevated Plus Maze
Activity in an elevated plus-maze apparatus with two open and two closed arms was measured over a 5-min period. The frequency and time spent in the open arms and closed arms were measured using the AnyMaze tracking system.
Open Field
Activity in an open field was measured over a 10-min period with AnyMaze. Time spent in margin and center, total distance, and mean speed were measured during the session.
Primary Cortical Neuron Culture
Embryonic brains were prepared from fetal Sprague-Dawley rats after 18 days of gestation and mechanically triturated. Dissociated cells were plated on six-well plates (0.6 million cells per well) in the plating medium consisting of Neurobasal media supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 1 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 2% Pen-Strep and 2% B27 supplement. We stopped proliferation of non-neuronal cells by replacing with plating medium lacking fetal serum the next day (DIV 1). Cultures were fed once a week with plating medium lacking fetal serum and maintained at 37°C in a humidified 5% CO2 atmosphere.
Construction, Preparation, and Infusion of Recombinant Adeno-Associated Virus
Hairpin RNA designed to specifically target the coding region of BICC1 mRNA was identified using published criteria (Hommel et al, 2003). Two sets of oligonucleotides were synthesized (Integrated DNA Technologies, Coralville, IA) and ligated into the mU6pro region of a modified pAAV-MCS vector, pAAV-shRNA. The pAAV-shRNA plasmid co-expressed the shRNA under control of a U6 promoter, and enhanced GFP under control of an independent RNA polymerase II promoter. Knockdown of BICC1 mRNA was verified in human embryonic kidney 293 (HEK293) cells and primary neuron culture.
AAV-BICC1shRNA:
Sense: 5′-TTTGATGTGGGGATCTTTCTTGGATTTCTTCCTGTCAAAATCCAAGAAAGATCCCCACATATTTTT-3′
Antisense: 5′-CTAGAAAAATATGTGGGGATCTTTCTTGGATTTTGACAGGAAGAAATCCAAGAAAGATCCCCACATC-3′
AAV-Scr:
Sense: 5′-TTTTTAGGTTATGGTGCGTAGTTGTTCCTTCCTGTCAGAACAACTACGCACCATAACCTAATTTTT-3′
Antisense: 5′-CTAGAAAAATTAGGTTATGGTGCGTAGTTGTTCTGACAGGAAGGAACAACTACGCACCATAACCTAA-3′
HEK293 cells were transfected with the AAV plasmid, along with the RC and helper plasmids, using a CaCl2 method. Seventy-two hours after transfection, cells were harvested and lysed, then purified using HiTrap heparin HP affinity columns (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Under a mixture of ketamine (100 mg/kg) and xylazine (6 mg/kg) anesthesia, virus was delivered bilaterally to rat hippocampus at a rate of 0.1 μl/min, followed by 3 min of rest to allow diffusion of the virus from the injector tip. The coordinates for the hippocampus were relative to bregma (−4.3, ±2 and −4.2 mm). After surgery, rats were given carprofen (5 mg/kg) as an analgesic and 5 ml of 0.9% saline to prevent fluid loss. Behavioral testing commenced 2 weeks following viral injection.
Statistical Analysis
Data were analyzed using two-tailed Student’s t-tests (for two-group comparisons) or ANOVAs (for multiple-group comparisons; SPSS), and differences were considered significant if p<0.05. Post hoc analyses were carried out using Fisher’s LSD test.
For postmortem studies, two-tailed paired t-tests were used to compare MDD to matched healthy controls, and differences were considered significant if p<0.05. In addition, ANCOVA was used to compare gene expression levels in MDD and psychiatrically healthy controls, and differences were considered significant if p<0.05. In the initial analysis, diagnostic status was the fixed factor, and age, PMI (postmortem interval), pH, and sex were included as covariates. In the reported analysis, we used diagnostic group as the main effect, and then included only those covariates that were significant in the analysis. Finally, to assess the effect of medication, we performed post hoc t-tests on pair differences in gene expression.
Outliers were removed from experiments if they were >2 SDs outside of the mean.
RESULTS
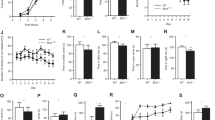
We assessed BICC1 mRNA levels in postmortem dlPFC and the DG and CA1 regions of the hippocampus in depressed subjects and matched controls (Figure 1). We observed a significant 1.49-fold increase of BICC1 mRNA in the dlPFC of MDD subjects relative to matched controls (t(36)=2.173, p<0.05). In the DG, we observed a significant 14.03-fold increase of BICC1 mRNA in MDD subjects relative to matched controls (t(6)=3.583, p<0.02). However, in the CA1 region, there was no significant regulation of BICC1 mRNA in MDD subjects. In each of these areas, covariate analyses showed no significant effect of age, sex, PMI, or pH, and post hoc t-test analysis showed no significant effect of medication. Further, in these unpaired ANCOVA models, the significance of the group effects remained consistent with that reported in the paired analysis (dlPFC: F(1,69)=8.093, p=0.006; DG: F(1,12)=4.820, p=0.049; CA1: F(1,12)=0.544, p=0.475). These data suggest that increased levels of BICC1 in the dlPFC and DG are associated with MDD and are consistent with the anatomical and functional changes that have previously been observed (Bermingham et al, 2012), where hippocampal size and dlPFC function were both affected by the BICC1 SNP.
BICC1 mRNA expression is increased in the dlPFC and DG of human MDD subjects. Results are mean±SEM fold change of BICC1 mRNA in punches taken from the dlPFC, DG, or CA1 of MDD patients or matched psychiatrically healthy controls (control N=37, MDD N=37 for dlPFC; control N=7, MDD N=7 for DG and CA1). Target mRNA levels have been normalized to GAPDH mRNA. Fold change of each MDD subject is calculated relative to its paired control; *p<0.05 controls vs MDD; two-tailed paired t-test.
To further assess BICC1 regulation in response to stress, we used the CUS model of depression in rats. Chronic stress is one of the better rodent models of depression, resulting in structural alterations as well as behavioral anhedonia, a core symptom of depression (Willner, 1997). Following 21 days of CUS, rats showed significant upregulation of BICC1 mRNA in both the hippocampus and PFC (Figure 2a). In the PFC, BICC1 mRNA showed a 1.42-fold increase in CUS animals compared to levels of expression in control animals (t(14)=2.773, p<0.05). In the hippocampus, BICC1 mRNA showed a 1.66-fold increase in CUS animals relative to levels of expression in control animals (t(12)=4.411, p<0.001). Furthermore, this effect appears to be dependent on chronic stress, as 1 day of mild stress (using the same stressors as the final day of the 21-day CUS paradigm; Figure 2b) or 2-h immobilization stress (Figure 2c) did not significantly regulate BICC1 in the PFC or hippocampus (p>0.05). Taken together, these data are consistent with the human postmortem results and show that chronic stress regulates BICC1 expression in the PFC and hippocampus.
Chronic unpredictable stress increases BICC1 in rat PFC and hippocampus. (a) Rats were subjected to 21 days of CUS or regular handling (Ctrl) and killed 4 h following the final stressor. Results are mean±SEM fold change BICC1 mRNA (Ctrl PFC n=8; CUS PFC n=8; Ctrl hippocampus n=7; and CUS hippocampus n=7) relative to control. (b) Rats received 1 day of mild stress (mild stress:1 day; identical to the final day of stressors used in the 21-day CUS protocol) or handling (Ctrl) then killed 4 h later. Results are mean±SEM fold change BICC1 mRNA (Ctrl PFC n=6; 1-day mild stress PFC n=6; Ctrl hippocampus n=6; and 1-day mild stress hippocampus n=5) relative to control. (c) Rats received 2-h immobilization stress (Imm stress: 2 h) or handling (Ctrl) then killed 4 h later. Results are mean±SEM fold change BICC1 mRNA (Ctrl PFC n=6; Imm PFC n=6; Ctrl hippocampus n=8; and Imm hippocampus, n=8) relative to control; *p<0.05 relative to Ctrl.
We also explored whether BICC1 may be involved in the actions of antidepressant treatments. Beurel et al (2011) showed that glycogen synthase kinase 3 (GSK3) is required for the rapid antidepressant effects of ketamine, and because both BICC1 and GSK3 are involved in Wnt signaling, we hypothesized that BICC1 may also play a role in mediating rapid antidepressant action. To test this hypothesis, we killed rats 2 h after administration of ketamine (10 mg/kg), a dose that has previously been shown to provide rapid antidepressant effects in rats (Li et al, 2010), then assessed BICC1 mRNA levels in the PFC and hippocampus. In the PFC, ketamine administration resulted in a significant 27% decrease relative to controls (t(14)=2.784, p<0.05; Figure 3a). In the hippocampus, ketamine administration produced a highly significant 90% decrease relative to controls (t(12)=6.813, p<0.0001; Figure 3a). We found that CUS-treated rats also showed a similar decrease in BICC1 expression following ketamine treatment (Supplementary Figure S1). Interestingly, rats chronically given fluoxetine, a selective serotonin reuptake inhibitor, do not show an effect on levels of BICC1 mRNA, suggesting that BICC1 regulation is highly dependent on the type of antidepressant (Figure 3b). This dichotomy in signaling pathways between rapid-acting antidepressants and SSRIs is consistent with previous work showing that mTORC1 signaling is recruited for the antidepressant actions of ketamine, but not chronic fluoxetine treatment (Li et al, 2010). In addition, ketamine increases glutamate (Moghaddam et al, 1997), which could cause neuronal depolarization and subsequently, decreased BICC1.
BICC1 mRNA is downregulated by acute ketamine but not by chronic fluoxetine. (a) Rats were injected with saline (Sal) or 10 mg/kg of ketamine (Ket), then killed 2 h later. Results are mean (±SEM) fold change for BICC1 mRNA expression (Sal PFC n=8; Ket PFC n=8; Sal hippocampus n=7; and Ket hippocampus n=7); *p<0.05 relative to Sal group. (b) Rats were injected with saline (Sal) or 5 mg/kg of fluoxetine (Fluox) for 21 days then killed 24 h later. Results are mean (±SEM) fold change for BICC1 mRNA expression in the hippocampus (Sal n=6; Fluox n=6).
We next used primary embryonic neuronal cultures to further explore the mechanism by which ketamine rapidly decreases BICC1 mRNA. We confirmed that incubation with a relatively low concentration of ketamine (500 nM) for 20 min decreases BICC1 mRNA expression (t(22)=2.977; p<0.01; Figure 4a). Further, we found that BICC1 mRNA is activity-regulated: specifically, we saw a significant decrease in BICC1 mRNA after incubation of primary neurons with a depolarizing concentration of KCl (40 mM; t(12)=2.264; p<0.05; Figure 4b). Conversely, we saw a significant increase in BICC1 mRNA expression after incubation with 1 μM TTX, which blocks voltage-gated sodium channels (t(10)=2.989; p<0.05; Figure 4c). In the future, it will be important to verify this in other culture systems, including adult primary neuronal cultures.
BICC1 mRNA is activity-regulated. (a) Primary cortical neuron cultures were treated on DIV 3 with either PBS or 500 nM ketamine for 20 min. Results are mean (±SEM) fold change for BICC1 mRNA expression (PBS, n=12; 500 nM, n=12). (b) Primary cortical neuron cultures were treated on DIV 3 with either PBS or 40 mM KCl for 20 min. Results are mean (±SEM) fold change for BICC1 mRNA expression (PBS, n=7; 40 mM, n=7). (c) Primary cortical neuron cultures were treated on DIV 3 with either PBS or 1 μM TTX for 20 min. Results are mean (±SEM) fold change for BICC1 mRNA expression (PBS, n=6; 1 μM, n=6); *p<0.05 relative to PBS group.
To further examine the functional effect of decreased BICC1, notably the decreased mRNA expression caused by ketamine, we utilized an shRNA strategy in combination with an AAV2 vector to achieve targeted knockdown of BICC1 expression. We chose to examine BICC1 knockdown in the hippocampus since this is where we observed the most dramatic decrease with ketamine. We first tested our AAV-BICC1shRNA in primary neuronal culture to verify knockdown and observed a 38% decrease in BICC1 expression relative to scrambled shRNA control (AAV-Scr; Supplementary Figure S2). We bilaterally injected either the AAV-BICC1shRNA or AAV-Scr into the rat hippocampus, specifically aiming for the DG (Figure 5a), where we had previously observed the highest BICC1 expression levels in depressed subjects. After a 2-week recovery period to allow for expression of AAV-BICC1shRNA, rats were exposed to CUS for 3 weeks, then tested for anhedonia, a core symptom of depression, using the sucrose preference test (Figure 5b). Two-factor ANOVA revealed a significant main effect of shRNA (F(1,28)=7.595; p<0.05) and a significant shRNA x stress interaction (F(1,28)=10.28; p<0.01), but no significant main effect of stress. Post hoc analysis showed that CUS/AAV-Scr showed significantly decreased sucrose preference relative to CTRL (p<0.05), but AAV-BICC1shRNA injection in CUS animals was able to rescue this effect (p<0.05). Importantly, there was no significant difference between CUS/AAV-BICC1shRNA as compared to either of the CTRL groups, suggesting that knockdown of BICC1 was sufficient for rescue of CUS-induced anhedonia. We also tested unstressed AAV-BICC1shRNA-injected animals to assess whether BICC1 knockdown had any effects on baseline behavior. We found that there was no significant difference between AAV-BICC1shRNA and AAV-Scr injected rats in the novelty suppressed feeding, forced swim, elevated plus-maze, or open-field tests, suggesting that BICC1 knockdown alone does not have a baseline effect on these behavioral modalities (Supplementary Figure S3a–d). Importantly, there was no difference in locomotor activity between groups (Supplementary Figure S3e and f).
Knockdown of BICC1 rescues depressive behavior following chronic unpredictable stress. (a) Representative injection site in the DG of the dorsal hippocampus, showing GFP expression in infected cells. DG, dentate gyrus; CA1, CA1 area of the hippocampus; CA3, CA3 area of the hippocampus. (b) Rats were given intrahippocampal injections of AAV-scrambled or AAV-BICC1shRNA, exposed to CUS, then subjected to behavioral testing. Results are mean (±SEM) percent preference in the sucrose preference test (Ctrl/AAV-scrambled n=8; Ctrl/AAV-BICC1shRNA n=8; CUS/AAV-Scrambled n=8; and CUS/AAV-BICC1shRNA n=8); *p<0.05 relative to Ctrl/AAV-scrambled group.
DISCUSSION
Taken together, the results demonstrate that levels of BICC1 are increased in the PFC and hippocampus of postmortem MDD subjects and in a rodent chronic stress model of depression. These findings are consistent with the possibility that the protective allele in the BICC1 SNP observed in the GWAS study may functionally decrease BICC1 levels to exert its protective effect. Further studies will be necessary to confirm this hypothesis.
In addition, using a combination of both in vivo and in vitro methods, we show a novel phenomenon of rapid BICC1 downregulation in response to acute ketamine treatment in an activity-dependent manner. BICC1 mRNA has a half-life of 11 h (Sharova et al, 2009), suggesting that the rapid decrease that we observe (within 2 h in vivo and 20 min in vitro) in BICC1 mRNA following ketamine and KCl treatment requires active degradation, as opposed to passive decay. Previous work has shown that BICC1 directly interacts with NOT3/5 to recruit the CCR4-NOT deadenylase complex to its target RNAs, including its own RNA (Chicoine et al, 2007), which provides a potential mechanism by which BICC1 could be rapidly degraded. However, the exact mechanisms for the rapid and robust downregulation of BICC1 mRNA in response to ketamine and KCl treatment will require additional studies. The activity-dependent downregulation of BICC1 is particularly interesting as genes showing activity-induced downregulation are relatively rare: they are an order of magnitude lower in abundance than genes showing activity-dependent upregulation (Hevroni et al, 1998; West et al, 2002). Here, we also show that the downregulation of BICC1 is functionally relevant, as knockdown of BICC1 in the hippocampus is sufficient to rescue anhedonic behavior in CUS-treated rats.
Previous studies have shown that BICC1 binds RNA via its K homology (KH) domains; these domains are highly conserved across species (Bouvrette et al, 2008), and mutation of these domains interferes with functional RNA-binding activity (Mahone et al, 1995). Although we know that much of BICC1’s function stems from regulation of mRNA stability and translation via modulation of mRNA adenylation through the KH domains, its exact RNA targets are just beginning to be elucidated (Chicoine et al, 2007; Mahone et al, 1995). Recent studies have provided some promising candidates that could elucidate the downstream mechanisms by which BICC1 exerts its effects on depressive behavior (Supplementary Figure S4). For example, overexpression of BICC1 resulted in co-immunoprecipitation with par-1, Cp190, Cip4, Klp10A, RhoGAP18B, and CG17293, which are all suggested or known to have roles in cytoskeletal regulation (Chicoine et al, 2007) and could potentially affect synaptic plasticity, which is disrupted by chronic stress and depression (Li et al, 2011; Liu and Aghajanian, 2008). Previous work has also shown that BICC1, via its SAM domain, can block Wnt signaling (Kraus et al, 2012) by uncoupling disheveled-2 signaling from the canonical Wnt pathway (Maisonneuve et al, 2009). The Wnt pathway has been implicated in the pathophysiology of depression (Voleti et al, 2011), suggesting another potential mechanism by which BICC1 could regulate MDD. Characterization of the exact RNA targets of BICC1 and studies of Wnt signaling will lead to a better understanding of the potential downstream mechanisms influenced by BICC1 that are relevant to stress and depression.
These results highlight a new approach to elucidating the molecular mechanisms underlying psychiatric disease by utilizing information derived from large GWAS studies. Taken together, these findings provide a potential novel molecular target both for the treatment of MDD and for gaining a further understanding of the molecular mechanisms underlying depression.
FUNDING AND DISCLOSURE
This work was supported by NIMH R37MH45481 (RSD), NIMH R01MH93897 (RSD), NIMH F32MH98513 (KTO), and NIGMS P30GM103328 (CAS), the State of Connecticut, and Yale University. RSD currently receives investigator-initiated research support from Forest, Lilly, and Sunovion and in 2012–2014 served as a consultant in the areas of target identification and validation and new compound development to Johnson & Johnson, Bristol-Myers Squibb, Taisho, Lundbeck, Asubio, and Sunovion. DAL currently receives investigator-initiated research support from Bristol-Myers Squibb and Pfizer and in 2012–2014 served as a consultant in the areas of target identification and validation and new compound development to Autifony, Bristol-Myers Squibb, Concert Pharmaceuticals, and Sunovion. KTO currently receives compensation from Bayer HealthCare Pharmaceuticals for professional service unrelated to the work presented in this manuscript.
References
Bermingham R, Carballedo A, Lisiecka D, Fagan A, Morris D, Fahey C et al (2012). Effect of genetic variant in BICC1 on functional and structural brain changes in depression. Neuropsychopharmacology 37: 2855–2862.
Beurel E, Song L, Jope RS (2011). Inhibition of glycogen synthase kinase-3 is necessary for the rapid antidepressant effect of ketamine in mice. Mol Psychiatry 16: 1068–1070.
Bouvrette DJ, Price SJ, Bryda EC (2008). K homology domains of the mouse polycystic kidney disease-related protein, bicaudal-C (Bicc1), mediate RNA binding in vitro. Nephron Exp Nephrol 108: e27–e34.
Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P (2007). Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell 13: 691–704.
Drevets WC, Price JL, Simpson JR, Todd RD, Reich T, Vannier M et al (1997). Subgenual prefrontal cortex abnormalities in mood disorders. Nature 386: 824–827.
Duric V, Banasr M, Stockmeier CA, Simen AA, Newton SS, Overholser JC et al (2013). Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. Int J Neuropsychopharmacol 16: 69–82.
Guay-Woodford LM (2003). Murine models of polycystic kidney disease: molecular and therapeutic insights. Am J Physiol Renal Physiol 285: F1034–F1049.
Hevroni D, Rattner A, Bundman M, Lederfein D, Gabarah A, Mangelus M et al (1998). Hippocampal plasticity involves extensive gene induction and multiple cellular mechanisms. J Mol Neurosci 10: 75–98.
Hommel JD, Sears RM, Georgescu D, Simmons DL, Dileone RJ (2003). Local gene knockdown in the brain using viral-mediated RNA interference. Nat Med 9: 1539–1544.
Izquierdo A, Wellman CL, Holmes A (2006). Brief uncontrollable stress causes dendritic retraction in infralimbic cortex and resistance to fear extinction in mice. J Neurosci 26: 5733–5738.
Kang HJ, Adams DH, Simen A, Simen BB, Rajkowska G, Stockmeier CA et al (2007). Gene expression profiling in postmortem prefrontal cortex of major depressive disorder. J Neurosci 27: 13329–13340.
Kang HJ, Voleti B, Hajszan T, Rajkowska G, Stockmeier CA, Licznerski P et al (2012). Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat Med 18: 1413–1417.
Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE (2005). Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 62: 617–627.
Kraus MR, Clauin S, Pfister Y, Di Maio M, Ulinski T, Constam D et al (2012). Two mutations in human BICC1 resulting in Wnt pathway hyperactivity associated with cystic renal dysplasia. Hum Mutat 33: 86–90.
Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K et al (2010). Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry 167: 949–957.
Li N, Lee B, Liu R-J, Banasr M, Dwyer JM, Iwata M et al (2010). mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science 329: 959–964.
Li N, Liu RJ, Dwyer JM, Banasr M, Lee B, Son H et al (2011). Glutamate N-methyl-D-aspartate receptor antagonists rapidly reverse behavioral and synaptic deficits caused by chronic stress exposure. Biol Psychiatry 69: 754–761.
Liu RJ, Aghajanian GK (2008). Stress blunts serotonin- and hypocretin-evoked EPSCs in prefrontal cortex: role of corticosterone-mediated apical dendritic atrophy. Proc Natl Acad Sci USA 105: 359–364.
Mahone M, Saffman EE, Lasko PF (1995). Localized bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. EMBO J 14: 2043–2055.
Maisonneuve C, Guilleret I, Vick P, Weber T, Andre P, Beyer T et al (2009). Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development 136: 3019–3030.
Moghaddam B, Adams B, Verma A, Daly D (1997). Activation of glutamatergic neurotransmission by ketamine: a novel step in the pathway from NMDA receptor blockade to dopaminergic and cognitive disruptions associated with the prefrontal cortex. J Neurosci 17: 2921–2927.
Radley JJ, Rocher AB, Miller M, Janssen WG, Liston C, Hof PR et al (2006). Repeated stress induces dendritic spine loss in the rat medial prefrontal cortex. Cereb Cortex 16: 313–320.
Radley JJ, Sisti HM, Hao J, Rocher AB, McCall T, Hof PR et al (2004). Chronic behavioral stress induces apical dendritic reorganization in pyramidal neurons of the medial prefrontal cortex. Neuroscience 125: 1–6.
Rajkowska G, Miguel-Hidalgo JJ, Wei J, Dilley G, Pittman SD, Meltzer HY et al (1999). Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol Psychiatry 45: 1085–1098.
Shansky RM, Hamo C, Hof PR, McEwen BS, Morrison JH (2009). Stress-induced dendritic remodeling in the prefrontal cortex is circuit specific. Cereb Cortex 19: 2479–2484.
Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MSH (2009). Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res 16: 45–58.
Sibille E, Morris HM, Kota RS, Lewis DA (2011). GABA-related transcripts in the dorsolateral prefrontal cortex in mood disorders. Int J Neuropsychopharmacol 14: 721–734.
Snee MJ, Macdonald PM (2009). Bicaudal C and trailer hitch have similar roles in gurken mRNA localization and cytoskeletal organization. Dev Biol 328: 434–444.
Voleti B, Tanis KQ, Newton SS, Duman RS (2011). Analysis of target genes regulated by chronic electroconvulsive therapy reveals role for Fzd6 in depression. Biol Psychiatry 71: 51–58.
West AE, Griffith EC, Greenberg ME (2002). Regulation of transcription factors by neuronal activity. Nat Rev Neurosci 3: 921–931.
Willner P (1997). Validity, reliability and utility of the chronic mild stress model of depression: a 10-year review and evaluation. Psychopharmacology (Berl) 134: 319–329.
Willner P (2005). Chronic mild stress (CMS) revisited: consistency and behavioural-neurobiological concordance in the effects of CMS. Neuropsychobiology 52: 90–110.
Acknowledgements
We are grateful to the families consenting to donate brain tissue and be interviewed for the human tissue samples, and to the Cuyahoga County Medical Examiner’s Office for their assistance. We thank Dr Grazyna Rajkowska for identification of anatomically comparable regions of dlPFC and Dr Vanja Duric for technical assistance with preparation of the postmortem samples. We thank Dr Mounira Banasr and Ashley Lepack for preparation of primary neuronal cultures and for technical assistance with the fluoxetine-treated rat samples. We also thank Dr Ralph J DiLeone for providing us with the pAAV-GFP-shRNA backbone plasmid.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Neuropsychopharmacology website
Supplementary information
Rights and permissions
About this article
Cite this article
Ota, K., Andres, W., Lewis, D. et al. BICC1 Expression is Elevated in Depressed Subjects and Contributes to Depressive Behavior in Rodents. Neuropsychopharmacol 40, 711–718 (2015). https://doi.org/10.1038/npp.2014.227
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2014.227
This article is cited by
-
Predicting the diagnosis of various mental disorders in a mixed cohort using blood-based multi-protein model: a machine learning approach
European Archives of Psychiatry and Clinical Neuroscience (2023)