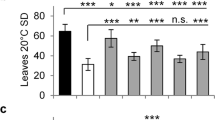
Abstract
Increasing global temperatures have an impact on flowering, and the underlying mechanisms are just beginning to be unravelled1,2. Elevated temperatures can induce flowering, and different mechanisms that involve either activation or de-repression of FLOWERING LOCUS T (FT) by transcription factor PHYTOCHROME INTERACTING FACTOR 4 (PIF4) or the FLOWERING LOCUS M (FLM)–SHORT VEGETATIVE PHASE (SVP) complex, respectively, have been suggested to be involved3–6. Thermosensitivity in flowering has been mapped to FLM5, which encodes a floral repressor7,8. FLM undergoes alternative splicing8 and it has been suggested that temperature-dependent alternative splicing leads to differential accumulation of the FLM-β and FLM-δ transcripts, encoding proteins with antagonistic effects, and that their ratio determines floral transition4. Here we show that high temperatures downregulate FLM expression by alternative splicing coupled with nonsense-mediated mRNA decay (AS-NMD). We identify thermosensitive splice sites in FLM and show that the primary effect of temperature is explained by an increase in NMD target transcripts. We also show that flm is epistatic to pif4, which suggests that most of the PIF4 effects are FLM dependent. Our findings suggest a model in which the loss of the floral repressor FLM occurs through mRNA degradation in response to elevated temperatures, signifying a role for AS-NMD in conferring environmental responses in plants.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fitter, A. H. & Fitter, R. S. Rapid changes in flowering time in British plants. Science 296, 1689–1691 (2002).
Wigge, P. A. Ambient temperature signalling in plants. Curr. Opin. Plant Biol. 16, 661–666 (2013).
Kumar, S. V. et al. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242–245 (2012).
Pose, D. et al. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417 (2013).
Balasubramanian, S., Sureshkumar, S., Lempe, J. & Weigel, D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2, e106 (2006).
Lee, J. H. et al. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628–632 (2013).
Scortecci, K., Michaels, S. D. & Amasino, R. M. Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol. Biol. 52, 915–922 (2003).
Scortecci, K. C., Michaels, S. D. & Amasino, R. M. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26, 229–236 (2001).
Kalyna, M. et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40, 2454–2469 (2012).
Drechsel, G. et al. Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25, 3726–3742 (2013).
Reddy, A. S., Marquez, Y., Kalyna, M. & Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 25, 3657–3683 (2013).
Severing, E. I. et al. Predicting the impact of alternative splicing on plant MADS domain protein function. PLoS ONE 7, e30524 (2012).
Arciga-Reyes, L., Wootton, L., Kieffer, M. & Davies, B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47, 480–489 (2006).
Hori, K. & Watanabe, Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 43, 530–540 (2005).
Ishigaki, Y., Li, X., Serin, G. & Maquat, L. E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617 (2001).
Lutz, U. et al. Modulation of ambient temperature-dependent flowering in Arabidopsis thaliana by natural variation of FLOWERING LOCUS M. PLoS Genet. 11, e1005588 (2015).
Sheldon, C. C. et al. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458 (1999).
Rosloski, S. M. et al. Functional analysis of splice variant expression of MADS AFFECTING FLOWERING 2 of Arabidopsis thaliana. Plant Mol. Biol. 81, 57–69 (2013).
Jiao, Y. & Meyerowitz, E. M. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 6, 419 (2010).
Werner, J. D. et al. Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc. Natl Acad. Sci. USA 102, 2460–2465 (2005).
Galvao, V. C., Collani, S., Horrer, D. & Schmid, M. Gibberellic acid signaling is required for ambient temperature-mediated induction of flowering in Arabidopsis thaliana. Plant J. 84, 949–962 (2015).
Peccarelli, M. & Kebaara, B. W. Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryot. Cell 13, 1126–1135 (2014).
Filichkin, S. A. et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58 (2010).
Barberan-Soler, S., Lambert, N. J. & Zahler, A. M. Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans. RNA 15, 1652–1660 (2009).
Kurihara, Y. et al. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl Acad. Sci. USA 106, 2453–2458 (2009).
Filichkin, S. A. et al. Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant 8, 207–227 (2015).
Gloggnitzer, J. et al. Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16, 376–390 (2014).
Ding, F. et al. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics 15, 431 (2014).
Chang, C. Y., Lin, W. D. & Tu, S. L. Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol. 165, 826–840 (2014).
Shikata, H. et al. Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl Acad. Sci. USA 111, 18781–6 (2014).
Leivar, P. et al. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20, 337–352 (2008).
de Lucas, M. et al. A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484 (2008).
Zhu, W. et al. Natural variation identifies ICARUS1, a universal gene required for cell proliferation and growth at high temperatures in Arabidopsis thaliana. PLoS Genet. 11, e1005085 (2015).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S. & Mullineaux, P. M. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832 (2000).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Acknowledgements
We thank C.A. Kraupner-Taylor for technical support, the Arabidopsis Biological Resource Centre (ABRC) and C. Fankhauser for seeds, H. Eimer and W. Zhu for providing some of the DNA and RNA samples used in this study, and J. Bowman, R. Burke, R. Clark, D. Smyth and W. Zhu for critical reading of the manuscript. This work is supported by Australian Research Council Discovery Projects DP110100964 and DP0987835 to S.S. and S.B., respectively, an ARC-Discovery Early Career Researcher Award (DE130100320) to C.T., an ARC Future Fellowship (FT100100377) to S.B. and funds from the Larkins Fellowship programme at Monash University to S.B.
Author information
Authors and Affiliations
Contributions
S.S. and S.B. designed the study. S.S. carried out most of the experiments. S.S., C.D., A.S., C.T. and S.B. analysed the data. S.S., C.D. and S.B. wrote the manuscript with input from all authors. S.B. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Different splice forms identified through Sanger sequencing. (XLSX 41 kb)
Supplementary Table 2
Effect of temperature on splicing at the FLM locus in Col-0. (XLSX 32 kb)
Supplementary Table 3
Actual counts and normalized counts from RNA-Seq. (XLSX 57 kb)
Supplementary Table 4
Flowering time measured as total leaf number in accessions that are potential knockouts for FLM-δ. (XLSX 48 kb)
Supplementary Table 5
Primers used in this study. (XLSX 39 kb)
Supplementary Table 6
NMD target genes, whose expression is analysed in Col-0 and upf 1-5 mutants. (XLSX 42 kb)
Rights and permissions
About this article
Cite this article
Sureshkumar, S., Dent, C., Seleznev, A. et al. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nature Plants 2, 16055 (2016). https://doi.org/10.1038/nplants.2016.55
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2016.55
This article is cited by
-
Nonsense-mediated mRNA decay pathway in plants under stress: general gene regulatory mechanism and advances
Planta (2024)
-
Mapping intron retention events contributing to complex traits using splice quantitative trait locus
Plant Methods (2023)
-
Fine-Tuning of the Grain Size by Alternative Splicing of GS3 in Rice
Rice (2022)
-
CRY2 interacts with CIS1 to regulate thermosensory flowering via FLM alternative splicing
Nature Communications (2022)
-
The splicing factor 1–FLOWERING LOCUS M module spatially regulates temperature-dependent flowering by modulating FLOWERING LOCUS T and LEAFY expression
Plant Cell Reports (2022)