Abstract
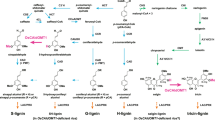
L-Phenylalanine ammonia-lyase (PAL) is the first enzyme in the biosynthesis of phenylpropanoid-derived plant compounds such as flavonoids, coumarins and the cell wall polymer lignin. The cell walls of grasses possess higher proportions of syringyl (S)-rich lignins and high levels of esterified coumaric acid compared with those of dicotyledonous plants, and PAL from grasses can also possess tyrosine ammonia-lyase (TAL) activity, the reason for which has remained unclear. Using phylogenetic, transcriptomic and in vitro biochemical analyses, we identified a single homotetrameric bifunctional ammonia-lyase (PTAL) among eight BdPAL enzymes in the model grass species Brachypodium distachyon. 13C isotope labelling experiments along with BdPTAL1-downregulation in transgenic plants showed that the TAL activity of BdPTAL1 can provide nearly half of the total lignin deposited in Brachypodium, with a preference for S-lignin and wall-bound coumarate biosynthesis, indicating that PTAL function is linked to the characteristic features of grass cell walls. Furthermore, isotope dilution experiments suggest that the pathways to lignin from L-phenylalanine and L-tyrosine are distinct beyond the formation of 4-coumarate, supporting the organization of lignin synthesis enzymes in one or more metabolons.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kenrick, P. & Crane, P. R. The origin and early evolution of plants on land. Nature 389, 33–39 (1997).
Bateman, R. M. et al. Early evolution of land plants: phylogeny, physiology, and ecology of the primary terrestrial radiation. Annu. Rev. Ecol. Syst. 29, 263–292 (1998).
Dixon, R. A. et al. The phenylpropanoid pathway and plant defence –a genomics perspective. Mol. Plant Pathol. 3, 371–390 (2002).
Maeda, H. & Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105 (2012).
Vanholme, R. et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341, 1103–1106 (2013).
Appert, C., Logemann, E., Hahlbrock, K., Schmid, J. & Amrhein, N. Structural and catalytic properties of the four phenylalanine ammonia-lyase isoenzymes from parsley (Petroselinum crispum Nym.). Eur. J. Biochem. 225, 491–499 (1994).
Kyndt, J. A., Meyer, T. E., Cusanovich, M. A. & Van Beeumen, J. J. Characterization of a bacterial tyrosine ammonia-lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett. 512, 240–244 (2002).
Cochrane, F. C., Davin, L. B. & Lewis, N. G. The Arabidopsis phenylalanine ammonia-lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry 65, 1557–1564 (2004).
Havir, E. A., Reid, P. D. & Marsh, H. V. L-phenylalanine ammonia-lyase (maize) evidence for a common catalytic site for L-phenylalanine and L-tyrosine. Plant Physiol. 48, 130–136 (1971).
Rosler, J., Krekel, F., Amrhein, N. & Schmid, J. Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 113, 175–179 (1997).
Jangaard, N. O. The characterization of phenylalanine ammonia-lyase from several plant species. Phytochemistry 13, 1765–1768 (1974).
Hsieh, L., Ma, G., Yang, C. & Lee, P. Cloning, expression, site-directed mutagenesis and immunolocalization of phenylalanine ammonia-lyase in Bambusa oldhamii. Phytochemistry 71, 1999–2009 (2010).
Brown, S. A., Wright, D. & Neish, A. C. Studies of lignin biosynthesis using isotopic carbon: VII.: The role of p-hydroxyphenylpyruvic acid. Can. J. Biochem. Physiol. 37, 25–34 (1959).
Higuchi, T., Ito, Y. & Kawamura, I. p-Hydroxyphenylpropane component of grass lignin and role of tyrosine-ammonia lyase in its formation. Phytochemistry 6, 875–881 (1967).
Louie, G. V. et al. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem. Biol. 13, 1327–1338 (2006).
Watts, K. T., Mijts, B. N., Lee, P. C., Manning, A. J. & Schmidt-Dannert, C. Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem. Biol. 13, 1317–1326 (2006).
Rétey, J. Discovery and role of methylidene imidazolone, a highly electrophilic prosthetic group. Biochim. Biophys. Acta 1647, 179–184 (2003).
MacDonald, M. J. & D'Cunha, G. B. A modern view of phenylalanine ammonia-lyase. Biochem. Cell Biol. 85, 273–282 (2007).
Cass, C. et al. Effects of phenylalanine ammonia lyase (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J. Exp. Bot. 66, 4317–4335 (2015).
Reichert, A., He, X. & Dixon, R. A. Phenylalanine ammonia-lyase (PAL) from tobacco (Nicotiana tabacum): characterization of the four tobacco PAL genes and active heterotetrameric enzymes. Biochem. J. 424, 233–242 (2009).
Hyun, M. W., Yun, Y. H., Kim, J. Y. & Kim, S. H. Fungal and plant phenylalanine ammonia-lyase. Mycobiology 39, 257–265 (2011).
Vogel, J. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 11, 301–307 (2008).
Harris, P. J. & Hartley, R. D. Detection of bound ferulic acid in cell walls of the Gramineae by ultraviolet fluorescence microscopy. Nature 259, 508–510 (1976).
Mann, D. G. J. et al. Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol. J. 10, 226–236 (2012).
Rasmussen, S. & Dixon, R. A. Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11, 1537–1551 (1999).
Achnine, L., Blancaflor, E. B., Rasmussen, S. & Dixon, R. A. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16, 3098–3109 (2004).
Bassard, J.-E. et al. Protein–protein and protein–membrane associations in the lignin pathway. Plant Cell 24, 4465–4482 (2012).
Winkel, B. S. J. Metabolic channeling in plants. Annu. Rev. Plant Biol. 55, 85–107 (2004).
Zhao, Q., & Dixon, R. A. Altering the cell wall and its impact on plant disease: from forage to bioenergy. Annu. Rev. Phytopathol. 52, 62–91 (2014).
Yamamoto, K., Nobayashi, N., Yoshitama, K., Teramoto, S. & Komamine, A. Isolation and purification of tyrosine hydroxylase from callus cultures of Portulaca grandiflora. Plant Cell Physiol. 42, 969–975 (2001).
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003).
Nuin, P. A. S., Wang, Z. & Tillier, E. R. M. The accuracy of several multiple sequence alignment programs for proteins. BMC Bioinform. 7, 471 (2006).
Scott, D. A., Hammond, P. M., Brearley, G. M. & Price, C. P. Identification by high-performance liquid chromatography of tyrosine ammonia-lyase activity in purified fractions of Phaseolus vulgaris phenylalanine ammonia-lyase. J. Chromatogr. B 573, 309–312 (1992).
Shevchenko, A., Tomas, H., Havli, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols 1, 2856–2860 (2006).
Rolando, C., Monties, B. & Lapierre, C. in Methods in Lignin Chemistry (eds Lin, S. Y. & Dence, C. W. ) 334–340 (Springer, 1992).
Broeckling, C. D. et al. Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J. Exp. Bot. 56, 323–336 (2005).
Rohde, A. et al. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16, 2749–2771 (2004).
Franke, R. et al. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30, 33–45 (2002).
Zuo, Y., Wang, C. & Zhan, J. Separation, characterization, and quantitation of benzoic and phenolic antioxidants in American cranberry fruit by GC–MS. J. Agricult. Food Chem. 50, 3789–3794 (2002).
Acknowledgements
We thank D. Huhman and the Noble Foundation Analytical Chemistry Facility for the analysis of soluble phenolic compounds, Dr X. Wang for assistance with protein purification, and Dr L. Gallego-Giraldo for critical reading of the manuscript. This work was supported by a Barrie Foundation Fellowship (to J.B.), The University of North Texas and the BioEnergy Science Center (Oak Ridge National Laboratory), a US Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Author information
Authors and Affiliations
Contributions
R.A.D. and J.B. conceived the study; J.B., J.S.-Y., F.C. and R.A.D. designed the experiments; J.B, J.S.-Y. and D.B. performed experimental work; J.B., J.S.-Y., D.B., B.V., F.C. and R.A.D. discussed and interpreted results; J.B. and R.A.D. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Barros, J., Serrani-Yarce, J., Chen, F. et al. Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nature Plants 2, 16050 (2016). https://doi.org/10.1038/nplants.2016.50
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2016.50
This article is cited by
-
Functional and kinetics of two efficient phenylalanine ammonia lyase from Pyrus bretschneideri
BMC Plant Biology (2023)
-
Transcriptomic and metabolic studies on the role of inorganic and organic iodine compounds in lettuce plants
Scientific Reports (2023)
-
Coordinated regulation of the entry and exit steps of aromatic amino acid biosynthesis supports the dual lignin pathway in grasses
Nature Communications (2023)
-
A comprehensive review of the classification, sources, biosynthesis, and biological properties of hydroxybenzoic and hydroxycinnamic acids
Phytochemistry Reviews (2023)
-
Integration of transcriptome and metabolome analyses reveals the role of OsSPL10 in rice defense against brown planthopper
Plant Cell Reports (2023)