Abstract
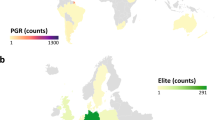
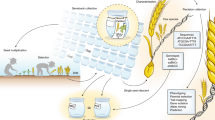
The 7.4 million plant accessions in gene banks are largely underutilized due to various resource constraints, but current genomic and analytic technologies are enabling us to mine this natural heritage. Here we report a proof-of-concept study to integrate genomic prediction into a broad germplasm evaluation process. First, a set of 962 biomass sorghum accessions were chosen as a reference set by germplasm curators. With high throughput genotyping-by-sequencing (GBS), we genetically characterized this reference set with 340,496 single nucleotide polymorphisms (SNPs). A set of 299 accessions was selected as the training set to represent the overall diversity of the reference set, and we phenotypically characterized the training set for biomass yield and other related traits. Cross-validation with multiple analytical methods using the data of this training set indicated high prediction accuracy for biomass yield. Empirical experiments with a 200-accession validation set chosen from the reference set confirmed high prediction accuracy. The potential to apply the prediction model to broader genetic contexts was also examined with an independent population. Detailed analyses on prediction reliability provided new insights into strategy optimization. The success of this project illustrates that a global, cost-effective strategy may be designed to assess the vast amount of valuable germplasm archived in 1,750 gene banks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Gerland, P. et al. World population stabilization unlikely this century. Science 346, 234–237 (2014).
Hoisington, D. et al. Plant genetic resources: what can they contribute toward increased crop productivity? Proc. Natl Acad. Sci. USA 96, 5937–5943 (1999).
Zamir, D. Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2, 983–989 (2001).
Tanksley, S. D. & McCouch, S. R. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066 (1997).
Houle, D., Govindaraju, D. R. & Omholt, S. Phenomics: the next challenge. Nat. Rev. Genet. 11, 855–866 (2010).
Morrell, P. L., Buckler, E. S. & Ross-Ibarra, J. Crop genomics: advances and applications. Nat. Rev. Genet. 13, 85–96 (2012).
Xu, S., Zhu, D. & Zhang, Q. Predicting hybrid performance in rice using genomic best linear unbiased prediction. Proc. Natl Acad. Sci. USA 111, 12456–12461 (2014).
Riedelsheimer, C. et al. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat. Genet. 44, 217–220 (2012).
de los Campos, G., Gianola, D. & Allison, D. B. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat. Rev. Genet. 11, 880–886 (2010).
Varshney, R. K. et al. Can genomics boost productivity of orphan crops? Nat. Biotech. 30, 1172–1176 (2012).
McCouch, S. et al. Agriculture: Feeding the future. Nature 499, 23–24 (2013).
Paterson, A. H. et al. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009).
Davey, J. W. et al. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 12, 499–510 (2011).
Elshire, R. J. et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6, e19379 (2011).
Thurber, C. S., Ma, J. M., Higgins, R. H. & Brown, P. J. Retrospective genomic analysis of sorghum adaptation to temperate-zone grain production. Genome Biol. 14, R68 (2013).
Morris, G. P. et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl Acad. Sci. USA 110, 453–458 (2012).
Darvasi, A. Experimental strategies for the genetic dissection of complex traits in animal models. Nat. Genet. 18, 19–24 (1998).
Morota, G. & Gianola, D. Kernel-based whole-genome prediction of complex traits: a review. Front. Genet. 5, 363 (2014).
Wray, N. R. et al. Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet. 14, 507–515 (2013).
Zaitlen, N. et al. Leveraging population admixture to characterize the heritability of complex traits. Nat. Genet. 46, 1356–1362 (2014).
Dekkers, J. C. M. Prediction of response to marker-assisted and genomic selection using selection index theory. J. Anim. Breed. Genet. 124, 331–341 (2007).
Tester, M. & Langridge, P. Breeding technologies to increase crop production in a changing world. Science 327, 818–822 (2010).
VanRaden, P. M. Efficient methods to compute genomic predictions. J. Dairy Sci. 91, 4414–4423 (2008).
Karaman, E., Cheng, H., Firat, M. Z., Garrick, D. J. & Fernando, R. L. An upper bound for accuracy of prediction using GBLUP. PLoS ONE 11, e0161054 (2016).
Harnessing the power of crop diversity to feed the future White Paper (DivSeek, 2014).
Meuwissen, T. H., Hayes, B. J. & Goddard, M. E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829 (2001).
Bernardo, R. & Yu, J. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 47, 1082–1090 (2007).
Heffner, E. L., Sorrells, M. E. & Jannink, J. L. Genomic selection for crop improvement. Crop Sci. 49, 1–12 (2009).
Crossa, J. et al. Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity 112, 48–60 (2014).
Spindel, J. et al. Genomic selection and association mapping in rice (Oryza sativa): effect of trait genetic architecture, training population composition, marker number and statistical model on accuracy of rice genomic selection in elite, tropical rice breeding lines. PLoS Genet. 11, e1004982 (2015).
Akdemir, D., Sanchez, J. I. & Jannink, J. L. Optimization of genomic selection training populations with a genetic algorithm. Genet. Sel. Evol. 47, 38 (2015).
Isidro, J. et al. Training set optimization under population structure in genomic selection. Theor. Appl. Genet. 128, 145–158 (2015).
Rincent, R. et al. Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (Zea mays L.). Genetics 192, 715–728 (2012).
Malosetti, M., Ribaut, J. M. & van Eeuwijk, F. A. The statistical analysis of multi-environment data: modeling genotype-by-environment interaction and its genetic basis. Front. Physiol. 4, 44 (2013).
Technow, F., Messina, C. D., Totir, L. R. & Cooper, M. Integrating crop growth models with whole genome prediction through approximate bayesian computation. PLoS ONE 10, e0130855 (2015).
Speed, D. & Balding, D. J. MultiBLUP: improved SNP-based prediction for complex traits. Genome Res. 24, 1550–1557 (2014).
Murray, M. G. & Thompson, W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325 (1980).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Bradbury, P. J. et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007).
Scheet, P. & Stephens, M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78, 629–644 (2006).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Liu, K. & Muse, S. V. Powermarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129 (2005).
Lipka, A. E. et al. GAPIT: Genome association and prediction integrated tool. Bioinformatics (2012).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013).
Endelman, J. B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4, 250–255 (2011).
de los Campos, G. et al. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182, 375–385 (2009).
Fernando, R. & Garrick, D. GenSel–User Manual for a Portfolio of Genomic Selection Related Analyses (Iowa State Univ., 2008); http://www.biomedcentral.com/content/supplementary/1471-2105-12-186-s1.pdf
Piepho, H. P. Ridge regression and extensions for genomewide selection in maize. Crop Sci. 49, 1165–1176 (2009).
Gianola, D., Fernando, R. L. & Stella, A. Genomic-assisted prediction of genetic value with semiparametric procedures. Genetics 173, 1761–1776 (2006).
Habier, D., Fernando, R., Kizilkaya, K. & Garrick, D. Extension of the bayesian alphabet for genomic selection. BMC Bioinformatics 12, 186 (2011).
Acknowledgements
This work was supported by the Agriculture and Food Research Initiative competitive grant (2011-03587) from the USDA National Institute of Food and Agriculture, by the National Science Foundation grant IOS-1238142, by the Kansas State University Center for Sorghum Improvement, by the Iowa State University Raymond F. Baker Center for Plant Breeding and by the Iowa State University Plant Science Institute. We appreciate K. Mayfield, L. Lambright and S. Staggenborg from Chromatin for conducting experiments at Lubbock, Texas.
Author information
Authors and Affiliations
Contributions
J.Y., M.L.W., G.A.P., T.T.T., P.S.S. and R.B. conceived and designed the experiments. X.Y., X.L., T.G., C.Z., Y.W., K.L.R., M.L.W. and J.Y. performed the experiments. X.Y., X.L. and C.Z. analysed the data. S.E.M., K.L.R., D.W., M.L.W., G.A.P., T.T.T., P.S.S. and R.B. contributed materials/analysis tools. X.Y., X.L., T.G. and J.Y. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-12 and Supplementary Tables 1-3. (PDF 2298 kb)
Supplementary Data Set
Trait data for the 299-accession training set, the 200-accession validation set, and the 45-accession validation set. (XLSX 50 kb)
Rights and permissions
About this article
Cite this article
Yu, X., Li, X., Guo, T. et al. Genomic prediction contributing to a promising global strategy to turbocharge gene banks. Nature Plants 2, 16150 (2016). https://doi.org/10.1038/nplants.2016.150
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2016.150
This article is cited by
-
Upcycling rice yield trial data using a weather-driven crop growth model
Communications Biology (2023)
-
Integrating genome-wide association study into genomic selection for the prediction of agronomic traits in rice (Oryza sativa L.)
Molecular Breeding (2023)
-
Target-oriented prioritization: targeted selection strategy by integrating organismal and molecular traits through predictive analytics in breeding
Genome Biology (2022)
-
Performance of Bayesian and BLUP alphabets for genomic prediction: analysis, comparison and results
Heredity (2022)
-
Genetic insights in pearl millet breeding in the genomic era: challenges and prospects
Plant Biotechnology Reports (2022)