Abstract
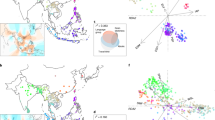
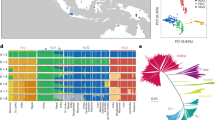
African rice (Oryza glaberrima) and African cultivation practices are said to have influenced emerging colonial plantation economies in the Americas1,2. However, the level of impact of African rice practices is difficult to establish because of limited written or botanical records2,3. Recent findings of O. glaberrima in rice fields of Suriname Maroons bear evidence of the high level of knowledge about rice among African slaves and their descendants, who consecrate it in ancestor rituals4,5. Here we establish the strong similarity, and hence likely origin, of the first extant New World landrace of O. glaberrima to landraces from the Upper Guinean forests in West Africa. We collected African rice from a Maroon market in Paramaribo, Suriname, propagated it, sequenced its genome6 and compared it with genomes of 109 accessions representing O. glaberrima diversity across West Africa. By analysing 1,649,769 single nucleotide polymorphisms (SNPs) in clustering analyses, the Suriname sample appears sister to an Ivory Coast landrace, and shows no evidence of introgression from Asian rice. Whereas the Dutch took most slaves from Ghana, Benin and Central Africa7, the diaries of slave ship captains record the purchase of food for provisions when sailing along the West African Coast8, offering one possible explanation for the patterns of genetic similarity. This study demonstrates the utility of genomics in understanding the largely unwritten histories of crop cultures of diaspora communities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wood, P. H. Black Majority: Negroes in Colonial South Carolina from 1670 through the Stono Rebellion (A.A. Knopf Inc., 1974).
Carney, J. A. Black Rice: The African Origins of Rice Cultivation in the Americas (Harvard Univ. Press, 2009).
Portères, R. Présence ancienne d'une variété cultivée d’Oryza glaberrima Steud. en Guyane Française. J. Agric. Trop. Bot. Appl. 2, 680 (1955).
Van Andel, T. R. African rice (Oryza glaberrima Steud.): lost crop of the enslaved Africans discovered in Suriname. Econ. Bot. 64, 1–10 (2010).
Van Andel, T. R., Van der Velden, A. & Reijers, M. The ‘Botanical Gardens of the Dispossessed’ revisited: richness and significance of Old World crops grown by Suriname Maroons. Genet. Resour. Crop Evol. 63, 695–710 (2016).
Gronau, I., Hubisz, M. J., Gulko, B., Danko, C. G. & Siepel, A. Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet. 43, 1031–1034 (2011).
Eltis, D. & Richardson, D. Atlas of the Transatlantic Slave Trade (Yale Univ. Press, 2010).
MCC Slave Voyage the Unity 1761-1763. http://eenigheid.slavenhandelmcc.nl/?lang=en (2013).
Carney, J. A. & Rosomoff, R. N. In the Shadow of Slavery: Africa's Botanical Legacy in the Atlantic World (Univ. California Press, 2009).
Voeks, R. A. & Rashford, J. African Ethnobotany in the Americas (Springer, 2012).
Meyer, R. S. et al. Domestication history and geographic adaptation inferred from a SNP map of African rice. Nat. Genet. 48, 1083–1088 (2016).
Molina, J. et al. Molecular evidence for a single evolutionary origin of domesticated rice. Proc. Natl. Acad. Sci. 108, 8351–8356 (2011).
Gilbert, E. in Rice: Global Networks and New Histories (ed. Bray, F. et al.) 212–228 (Cambridge Univ. Press, 2015).
Nuijten, E., van Treuren, R., Struik, P. C., Mokuwa, A. & Okry, F. Evidence for the emergence of new rice types of interspecific hybrid origin in West African farmers’ fields. PLoS ONE 4, e7335 (2009).
Sakagami, J. I., Joho, Y. & Sone, C. Complete submergence escape with shoot elongation ability by underwater photosynthesis in African rice, Oryza glaberrima Steud. F. Crop. Res. 152, 17–26 (2013).
Collinson, P. Of the introduction of rice and tar in our colonies. Gentleman's Magazine 278–280 (1766).
Vaillant, M. Milieu cultural et classification des variétés de riz des Guyanes française et hollandaise. Rev. Int. Bot. Appl. d'Agriculture Trop. 28, 520–529 (1948).
Herlein, J. D. Beschryvinge van de volk-plantinge Zuriname (Meindert Injema, 1718).
Stedman, J. G. Narrative of a Five Years’ Expedition, against the Revolted Negroes of Surinam 1790 (Johns Hopkins Univ. Press, 1988).
Price, S. Co-Wives and Calabashes (Univ. Michigan Press, 1993).
Geijskes, D. C. De landbouw bij de Bosnegers van de Marowijne. West-Indische Gids. 35, 135–153 (1955).
Baumgart, I. R., HilleRisLambers, D., Khodabaks, M. R. & Wildschut, J. Visit to rice growing sites on the Upper Suriname River between Nieuw Aurora and Abenaston, June 7-10 1998 (ADRON, 1998).
Wang, M. et al. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46, 982–988 (2014).
Mehra, P., Pandey, B. K. & Giri, J. Genome-wide DNA polymorphisms in low Phosphate tolerant and sensitive rice genotypes. Sci. Rep. 5, 13090 (2015).
Shah, S. M., Arif, M., Aslam, K., Shabir, G. & Thomson, M. J. Genetic diversity analysis of Pakistan rice (Oryza sativa) germplasm using multiplexed single nucleotide polymorphism markers. Genet. Resour. Crop. Evol. 63, 1113–1126 (2016).
Aflitos, S. A. et al. Introgression browser: high-throughput whole-genome SNP visualization. Plant J. 82, 174–182 (2015).
Fields-Black, E. L. Deep Roots: Rice Farmers in West Africa and the African Diaspora (Indiana Univ. Press, 2008).
Portères, R. in Origins of African Plant Domestication (ed. Harlan, J. R. ) 409–452 (Mouton Publishers, 1976).
Harlan, J. R. Origins of African Plant Domestication (Mouton Publishers, 1976).
Van Andel, T. R. et al. Local plant names reveal that enslaved Africans recognized substantial parts of the New World flora. Proc. Natl Acad. Sci. USA 111, E5346–E5353 (2014).
van Andel, T. R., Behari-Ramdas, J. A., Havinga, R. M. & Groenendijk, S. The medicinal plant trade in Suriname. Ethnobot. Res. Appl. 5, 351–372 (2007).
Taxon: Oryza glaberrima. Accession number AMD 20101002 Hortus Botanicus Amsterdam, the Netherlands; http://dehortus.gardenexplorer.org/taxon-3625.aspx
Joshi, N. A. & Fass, J. N. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files Version 1.33 (2011); https://github.com/najoshi/sickle
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Daneck, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2016).
Price, M. N., Dehal, P. S. & Arkin, A. P. Fasttree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Huerta-Cepas, J., Dopazo, J. & Gabaldón, T. ETE: a python environment for tree exploration. BMC Bioinformatics 11, 24 (2010).
Nychka, D., Furrer, R., Paige, J. & Sain, S. Fields: Tools for Spatial Data R package version 8.3-6 (2016).
Hijmans, R. J. Raster: Geographic Data Analysis and Modeling R package version 2.5-2 (2015); https://cran.r-project.org/package=raster
South, A. Rworldmap: a new R package for mapping global data. R. J. 3, 35–43 (2011).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2012); http://www.r-project.org
Freedman, A. H. et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 10, e1004016 (2014).
Acknowledgements
We would like to thank rice farmers in Jawjaw, Mundje Kreek, Mooytaki and Paramaribo for sharing their knowledge on African rice with us. T. Polimé and B. Poeketie facilitated fieldwork in Maroon communities. C.-R. Lee helped us with the TPS analysis. This research was funded by the Biosystematics group of Wageningen University, Naturalis Biodiversity Center (Leiden), as well as support from NSF Plant Genome to R.S.M. (IOS-1202803) and M.D.P. (IOS-1126971), a TKI-Horticulture Grant to M.E.S. and S.A.A., grants from the US National Science Foundation and the NYU Abu Dhabi Research Institute to J.M.F., and the AXA Chair in Genome Biology and Evolutionary Genomics to R.A.W.
Author information
Authors and Affiliations
Contributions
T.R.v.A. conducted the fieldwork in Suriname, and wrote the paper with contributions and input from all co-authors. T.R.v.A. and M.E.S. conceived and guided the research. R.M.H. maintained the living collection of Oryza glaberrima from Suriname used in this project. D.C. performed sequencing and assisted with data analysis. R.S.M. and J.M.F. performed alignments, SNP calling, and clustering analyses. S.A.A. and M.A.V. performed phylogenomic and TPS analysis. J.A.C. and H.M. provided background on the geographical and historical aspects of West African rice, slavery and Suriname. R.A.W. and M.D.P. helped to collect the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figure 1-5 (PDF 548 kb)
Supplementary Table 1
Background information on the 110 Oryza glaberrima samples used for analysis, including mean depth of sequencing coverage, accession codes, collection locations and clustering distance to Surinamese sample (suri_glab_SUR). (XLSX 24 kb)
Supplementary Table 2
Per cent heterozygosity of African rice accessions with additional country labels. (XLSX 51 kb)
Supplementary Table 3
Filtering of polymorphisms. Multi Nucleotide Polymorphisms (MNP) were always excluded. Heterozygous (HET) SNPs and regions with no coverage (NC) were tentatively excluded as well but their frequency was too high. (XLSX 8 kb)
Supplementary Table 4
Top 10 EIGENSTRAT principal components of the 110 Oryza glaberrima samples. (XLSX 21 kb)
Supplementary Table 5
Ethnolinguistic information on sample provenances. Ethnolinguistic classification of locations from which the samples were collected. (XLSX 14 kb)
Supplementary Table 6
Genetic distance between the O. glaberrima samples. Kruskal-Wallis multiple comparison tests. A Kruskal-Wallis test showed significant differences in genetic distances to the Surinamese sample by country (where country n>1) (Kruskal-Wallis chi-squared = 53.809, df = 11, p-value = 1.279e-07). Results reported are for the multiple comparison test following Kruskal-Wallis set to Bonferroni significance of alpha = 0.05 after correction for 66 tests (P<0.00075). (XLSX 44 kb)
Rights and permissions
About this article
Cite this article
van Andel, T., Meyer, R., Aflitos, S. et al. Tracing ancestor rice of Suriname Maroons back to its African origin. Nature Plants 2, 16149 (2016). https://doi.org/10.1038/nplants.2016.149
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2016.149
This article is cited by
-
Genetics and Genomics of African Rice (Oryza glaberrima Steud) Domestication
Rice (2021)
-
The legacy of traditional rice cultivation by descendants of Indian contract laborers in Suriname
Journal of Ethnobiology and Ethnomedicine (2021)
-
Whole-genome resequencing analysis of 20 Micro-pigs
Genes & Genomics (2020)
-
Agrobiodiversity and a sustainable food future
Nature Plants (2017)
-
Plant genomics: African origins of ‘black rice’
Nature Plants (2016)