Abstract
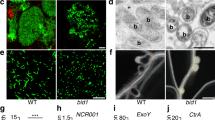
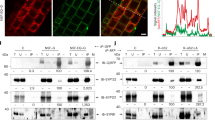
Many microbes interact with their hosts across a membrane interface, which is often distinct from existing membranes. Understanding how this interface acquires its identity has significant implications. In the symbiosis between legumes and rhizobia, the symbiosome encases the intracellular bacteria and receives host secretory proteins important for bacterial development. We show that the Medicago truncatula SYNTAXIN 132 (SYP132) gene undergoes alternative cleavage and polyadenylation during transcription, giving rise to two target-membrane soluble NSF attachment protein receptor (t-SNARE) isoforms. One of these isoforms, SYP132A, is induced during the symbiosis, is able to localize to the peribacteroid membrane, and is required for the maturation of symbiosomes into functional forms. The second isoform, SYP132C, has important functions unrelated to symbiosis. The SYP132A sequence is broadly found in flowering plants that form arbuscular mycorrhizal symbiosis, an ancestral mutualism between soil fungi and most land plants. SYP132A silencing severely inhibited arbuscule colonization, indicating that SYP132A is an ancient factor specifying plant–microbe interfaces.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ivanov, S., Fedorova, E. & Bisseling, T. Intracellular plant microbe associations: secretory pathways and the formation of perimicrobial compartments. Curr. Opin. Plant Biol. 13, 372–377 (2010).
Wang, D. et al. A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science 327, 1126–1129 (2010).
Zheng, H. et al. The syntaxin family of proteins in Arabidopsis: a new syntaxin homologue shows polymorphism between two ecotypes. J. Exp. Bot. 50, 915–924 (1999).
Catalano, C. M., Lane, W. S. & Sherrier, D. J. Biochemical characterization of symbiosome membrane proteins from Medicago truncatula root nodules. Electrophoresis 25, 519–531 (2004)
Catalano, C. et al. Medicago truncatula syntaxin SYP132 defines the symbiosome membrane and infection droplet membrane in root nodules. Planta 225, 541–550 (2007).
Limpens, E. et al. Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell 21, 2811–2828 (2009).
Benedito, V. A. et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 55, 504–513 (2008).
Elkon, R., Ugalde, A. P. & Agami, R. Alternative cleavage and polyadenylation: extent, regulation and function. Nature Rev. Genet. 14, 496–506 (2013).
Kalde, M. et al. The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl Acad. Sci. USA 104, 11850–11855 (2007).
Van de Velde, W. et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126 (2010).
Kondorosi, E., Mergaert, P. & Kereszt, A. A paradigm for endosymbiotic life cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu. Rev. Microbiol. 67, 611–628 (2013).
Delaux, P.-M. et al. Evolution of the plant-microbe symbiotic ‘toolkit’. Trends Plant Sci. 18, 298–304 (2013).
Ivanov, S. et al. Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc. Natl Acad. Sci. USA 109, 8316–8321 (2012).
Sinharoy, S. et al. The C2H2 transcription factor regulator of symbiosome differentiation represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula. Plant Cell 25, 3584–3601 (2013).
Collins, N. C. et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977 (2003).
Assaad, F. F. et al. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15, 5118–5129 (2004).
Meyer, D. et al. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 57, 986–999 (2009).
Kwon, C. et al. Co-option of a default secretory pathway for plant immune responses. Nature 451, 835–840 (2008).
Frazer, K. A. et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 (2004).
Curtis, M. D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
Helliwell, C. A. & Waterhouse, P. M. Constructs and methods for hairpin RNA-mediated gene silencing in plants. Meth. Enzymol. 392, 24–35 (2005).
Boisson-Dernier, A. et al. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14, 695–700 (2001).
Leong, S. A., Williams, P. H. & Ditta, G. S. Analysis of the 5′ regulatory region of the gene for δ-aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res. 13, 5965–5976 (1985).
Giovannetti, M. & Mosse, B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84, 489–500 (1980).
Kakar, K. et al. A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4, 18–30 (2008).
Altschul, S. F. et al. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999).
Acknowledgements
We are grateful to D. Hebert, S. Hazen and T. Baskin (University of Massachusetts, Amherst) for access to instruments and C. Haney (University of British Columbia, Vancouver) for modifying the pHellsGate8 vector with the mCherry reporter. We thank F. M. Ausubel (Massachusetts General Hospital, Boston), J. S. Griffitts (Brigham Young University, Provo), S. R. Long (Stanford University, Stanford) and A. Caicedo (University of Massachusetts, Amherst) for insightful discussions of the manuscript. This work was supported by the University of Massachusetts Amherst and USDA Hatch Grant (to D.W.).
Author information
Authors and Affiliations
Contributions
D.W., H.P., O.O., E.W. and B.W. designed the research; H.P., O.O., X.W. and X.Z. performed the experiments; D.W., C.S., B.W., X.W. performed alignments and evolutionary analysis; D.W., H.P., O.O. and C.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Pan, H., Oztas, O., Zhang, X. et al. A symbiotic SNARE protein generated by alternative termination of transcription. Nature Plants 2, 15197 (2016). https://doi.org/10.1038/nplants.2015.197
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2015.197
This article is cited by
-
Exploring the role of symbiotic modifier peptidases in the legume − rhizobium symbiosis
Archives of Microbiology (2024)
-
A signal peptide peptidase is required for ER-symbiosome proximal association and protein secretion
Nature Communications (2023)
-
Extensive membrane systems at the host–arbuscular mycorrhizal fungus interface
Nature Plants (2019)
-
Genome-wide atlas of alternative polyadenylation in the forage legume red clover
Scientific Reports (2018)
-
Nodule cysteine-rich peptides maintain a working balance during nitrogen-fixing symbiosis
Nature Plants (2017)