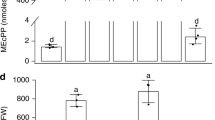
Abstract
The regulatory nucleotide guanosine 5′-diphosphate 3′-diphosphate (ppGpp), which was originally identified in Escherichia coli, controls the expression of a large gene set and many enzyme activities. The ppGpp-dependent control of cell activities is referred to as the stringent response. Recently, genes responsible for the synthesis and degradation of ppGpp have been identified not only in bacteria, but also in eukaryotes, including plants and animals, indicating that the stringent response is, unexpectedly, widely conserved. However, the exact function of the eukaryotic stringent response remains elusive. Here, we isolated an Arabidopsis mutant that overproduces ppGpp in chloroplasts. This mutant shows metabolite reduction, dwarf chloroplasts and significantly suppressed plastidial transcription and translation. Under nutrient-deficient conditions, the mutant shows more robust growth than the wild type. These results indicate that the ppGpp-dependent control of the organelle function is crucial for the systematic growth of host organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cashel, M., Gentry, D. R., Hernandez, V. J. & Vinella, D. in Escherichia coli and Salmonella: Cellular and Molecular Biology 2nd edn (ed. Neidhardt, F. C. ) 1458–1496 (ASM Press, 1996).
Cashel, M. The control of ribonucleic acid synthesis in Escherichia coli IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J. Biol. Chem. 244, 3133–3141 (1969).
Atkinson, G. C., Tenson, T. & Hauryliuk, V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 6, e23479 (2011).
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T. & Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nature Rev. Microbiol. 13, 298–309 (2015).
Potrykus, K. & Cashel, M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51 (2008).
Mechold, U., Potrykus, K., Murphy, H., Murakami, K. S. & Cashel, M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 41, 6175–6189 (2013).
Ross, W., Vrentas, C. E., Sanchez-Vazquez, P., Gaal, T. & Gourse, R. L. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 50, 420–429 (2013).
Zuo, Y., Wang, Y. & Steitz, T. A. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell 50, 430–436 (2013).
Milon, P. et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl Acad. Sci. USA 103, 13962–13967 (2006).
Rojas, A.-M., Ehrenberg, M. N., Andersson, S. G. & Kurland, C. ppGpp inhibition of elongation factors Tu, G and Ts during polypeptide synthesis. Mol. Gen. Genet. 197, 36–45 (1984).
Kanjee, U., Ogata, K. & Houry, W. A. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol. Microbiol. 85, 1029–1043 (2012).
Maisonneuve, E., Castro-Camargo, M. & Gerdes, K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150 (2013).
Masuda, S. in Advances in Photosynthesis – Fundamental Aspects (ed. Najafpour, M. ) 487–500 (In Tech, 2012).
Battesti, A. l. & Bouveret, E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62, 1048–1063 (2006).
Tozawa, Y. & Nomura, Y. Signalling by the global regulatory molecule ppGpp in bacteria and chloroplasts of land plants. Plant Biol. 13, 699–709 (2011).
van der Biezen, E. A., Sun, J., Coleman, M. J., Bibb, M. J. & Jones, J. D. Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc. Natl Acad. Sci. USA 97, 3747–3752 (2000).
Givens, R. M. et al. Inducible expression, enzymatic activity, and origin of higher plant homologues of bacterial RelA/SpoT stress proteins in Nicotiana tabacum. J. Biol. Chem. 279, 7495–7504 (2004).
Masuda, S. et al. The bacterial stringent response, conserved in chloroplasts, controls plant fertilization. Plant Cell Physiol. 49, 135–141 (2008).
Tozawa, Y. et al. Calcium-activated (p)ppGpp synthetase in chloroplasts of land plants. J. Biol. Chem. 282, 35536–35545 (2007).
Mizusawa, K., Masuda, S. & Ohta, H. Expression profiling of four RelA/SpoT-like proteins, homologues of bacterial stringent factors, in Arabidopsis thaliana. Planta 228, 553–562 (2008).
Ihara, Y., Ohta, H. & Masuda, S. A highly sensitive quantification method for the accumulation of alarmone ppGpp in Arabidopsis thaliana using UPLC-ESI-qMS/MS. J. Plant Res. 128, 511–518 (2015).
Takahashi, K., Kasai, K. & Ochi, K. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc. Natl Acad. Sci. USA 101, 4320–4324 (2004).
Zimorski, V., Ku, C., Martin, W. F. & Gould, S. B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 22, 38–48 (2014).
Liere, K., Weihe, A. & Börner, T. The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J. Plant Phys. 168, 1345–1360 (2011).
Sato, M. et al. Bacterial alarmone, guanosine 5′-diphosphate 3′-diphosphate (ppGpp), predominantly binds the β’ subunit of plastid-encoded plastid RNA polymerase in chloroplasts. ChemBioChem 10, 1227–1233 (2009).
Nagashima, A. et al. DNA microarray analysis of plastid gene expression in an Arabidopsis mutant deficient in a plastid transcription factor sigma, SIG2. Biosci. Biotechnol. Biochem. 68, 694–704 (2004).
Parry, M. A., Keys, A. J., Madgwick, P. J., Carmo-Silva, A. E. & Andralojc, P. J. Rubisco regulation: a role for inhibitors. J. Exp. Bot. 59, 1569–1580 (2008).
Coruzzi, G. M. & Zhou, L. Carbon and nitrogen sensing and signaling in plants: emerging “matrix effects”. Curr. Opin. Plant Biol. 4, 247–253 (2001).
Umena, Y., Kawakami, K., Shen, J.-R. & Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 (2011).
Krásný, L. & Gourse, R. L. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23, 4473–4483 (2004).
Liu, K. et al. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol. Cell 57, 735–749 (2015).
Miura, E. et al. The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19, 1313–1328 (2007).
Tiboni, O., Pasquale, G. & Ciferri, O. Purification of the elongation factors present in spinach chloroplasts. Eur. J. Biochem. 92, 471–477 (1978).
Rodermel, S. R., Abbott, M. S. & Bogorad, L. Nuclear-organelle interactions: nuclear antisense gene inhibits ribulose bisphosphate carboxylase enzyme levels in transformed tobacco plants. Cell 55, 673–681 (1988).
Bang, W. Y. et al. AtObgC, a plant ortholog of bacterial Obg, is a chloroplast-targeting GTPase essential for early embryogenesis. Plant Mol. Biol. 71, 379–390 (2009).
Chigri, F., Sippel, C., Kolb, M. & Vothknecht, U. C. Arabidopsis OBG-like GTPase (AtOBGL) is localized in chloroplasts and has an essential function in embryo development. Mol. Plant 2, 1373–1383 (2009).
Masuda, S., Tozawa, Y. & Ohta, H. Possible targets of magic spots in plant signalling. Plant Signal. Behav. 3, 1021–1023 (2008).
Chen, J. et al. AtObgC-AtRSH1 interaction may play a vital role in stress response signal transduction in Arabidopsis. Plant Physiol. Biochem. 74, 176–184 (2014).
Sulpice, R. et al. Starch as a major integrator in the regulation of plant growth. Proc. Natl Acad. Sci. USA 106, 10348–10353 (2009).
Ruan, Y.-L. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67 (2014).
Sheen, J. Master regulators in plant glucose signaling networks. J. Plant Biol. 57, 67–79 (2014).
Sun, D. et al. A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nature Struct. Mol. Biol. 17, 1188–1194 (2010).
Porra, R., Thompson, W. & Kriedemann, P. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 (1989).
Araya, T., Noguchi, K. & Terashima, I. Effect of nitrogen nutrition on the carbohydrate repression of photosynthesis in leaves of Phaseolus vulgaris L. J. Plant Res. 123, 371–379 (2010).
Ticconi, C. A., Delatorre, C. A. & Abel, S. Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol. 127, 963–972 (2001).
Yoshimoto, K. et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927 (2009).
Oikawa, A., Fujita, N., Horie, R., Saito, K. & Tawaraya, K. Solid-phase extraction for metabolomic analysis of high-salinity samples by capillary electrophoresis-mass spectrometry. J. Sep. Sci. 34, 1063–1068 (2011).
Spurr, A. R. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26, 31–43 (1969).
Acknowledgements
We thank R. Sasaki (RIKEN) , K. Yamamichi (Tokyo Institute of Technology) and K. Hori (Tokyo Institute of Technology) for excellent technical assistance, L. Kwok (Tokyo Institute of Technology) for critical reading of the manuscript, and T. Hisabori (Tokyo Institute of Technology), Y. Nagano (Saga University) and M. Ikeuchi (The University of Tokyo) for antibodies. We also thank the Arabidopsis Biological Resource Center (The Ohio State University) for providing mutant seeds. This work was supported by a Grants-in-Aid for Scientific Research on Innovative Areas from MEXT of Japan (No. 25120709) to S.M.
Author information
Authors and Affiliations
Contributions
M.M. and R.H. performed phenotypic analysis of mutants that includes gene expression analysis, immunoblot analysis, photosynthetic activity measurements and lipid analysis. Y.I. performed ppGpp quantification. R.S. analysed localization of GFP fusion proteins. R.H. and A.O. performed metabolome analysis, Y.K. and M.S. performed hormone quantification. All authors analysed data and discussed the results. S.M. designed the research. M.M. and S.M. wrote the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Maekawa, M., Honoki, R., Ihara, Y. et al. Impact of the plastidial stringent response in plant growth and stress responses. Nature Plants 1, 15167 (2015). https://doi.org/10.1038/nplants.2015.167
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2015.167
This article is cited by
-
Significance of the plastidial stringent response for plant growth on soil
Plant Growth Regulation (2024)
-
ppGpp accumulation reduces the expression of the global nitrogen homeostasis-modulating NtcA regulon by affecting 2-oxoglutarate levels
Communications Biology (2023)
-
Metabolic changes contributing to large biomass production in the Arabidopsis ppGpp-accumulating mutant under nitrogen deficiency
Planta (2022)
-
ppGpp functions as an alarmone in metazoa
Communications Biology (2020)
-
Significance of accumulation of the alarmone (p)ppGpp in chloroplasts for controlling photosynthesis and metabolite balance during nitrogen starvation in Arabidopsis
Photosynthesis Research (2018)