Abstract
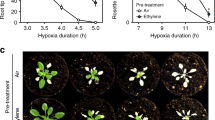
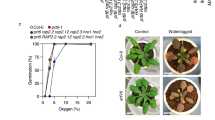
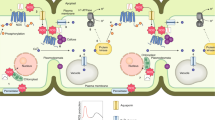
Plant survival is greatly impaired when oxygen levels are limiting, such as during flooding or when anatomical constraints limit oxygen diffusion. Oxygen sensing in Arabidopsis thaliana is mediated by Ethylene Responsive Factor (ERF)-VII transcription factors, which control a core set of hypoxia- and anoxia-responsive genes responsible for metabolic acclimation to low-oxygen conditions. Anoxic conditions also induce genes related to reactive oxygen species (ROS). Whether the oxygen-sensing machinery coordinates ROS production under anoxia has remained unclear. Here we show that a low-oxygen-responsive universal stress protein (USP), Hypoxia Responsive Universal Stress Protein 1 (HRU1), is induced by RAP2.12 (Related to Apetala 2.12), an ERF-VII protein, and modulates ROS production in Arabidopsis. We found that HRU1 is strongly induced by submergence, but that this induction is abolished in plants lacking RAP2.12. Mutation of HRU1 through transfer DNA (T-DNA) insertion alters hydrogen peroxide production, and reduces tolerance to submergence and anoxia. Yeast two-hybrid and bimolecular fluorescence complementation (BiFC) analyses reveal that HRU1 interacts with proteins that induce ROS production, the GTPase ROP2 and the NADPH oxidase RbohD, pointing to the existence of a low-oxygen-specific mechanism for the modulation of ROS levels. We propose that HRU1 coordinates oxygen sensing with ROS signalling under anoxic conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bailey-Serres, J. & Voesenek, L. A. Flooding stress: acclimations and genetic diversity. Annu. Rev. Plant Biol. 59, 313–339 (2008).
Kennedy, R. A., Rumpho, M. E. & Fox, T. C. Anaerobic metabolism in plants. Plant Physiol. 100, 1–6 (1992).
Ricard, B. et al. Plant metabolism under hypoxia and anoxia. Plant Physiol. Biochem. 32, 1–10 (1994).
Loreti, E., Poggi, A., Novi, G., Alpi, A. & Perata, P. A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 137, 1130–1138 (2005).
Mustroph, A. et al. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152, 1484–1500 (2010).
Lee, S. C. et al. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 190, 457–471 (2011).
Sasidharan, R. & Mustroph, A. Plant oxygen sensing is mediated by the N-end rule pathway: a milestone in plant anaerobiosis. Plant Cell. 23, 4173–4183 (2011).
Gibbs, D. J. et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418 (2011).
Licausi, F. et al. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422 (2011).
Chang, R., Jang, C. J., Branco-Price, C., Nghiem, P. & Bailey-Serres, J. Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis. Plant Mol. Biol. 78, 109–122 (2012).
Baxter-Burrel, A., Yang, Z., Springer, P. S. & Bailey-Serres, J. RopGAP4-dependent RopGTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296, 2026–2028 (2002).
Pucciariello, C., Parlanti, S., Banti, V., Novi, G. & Perata, P. Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol. 159, 184–196 (2012).
Bechtold, U. et al. Impact of chloroplastic- and extracellular-sourced ROS on high light-responsive gene expression in Arabidopsis. J. Exp. Bot. 59, 121–133 (2008).
Nachin, L., Nannmark, U. & Nyström, T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187, 6265–6272 (2005).
Kvint, K., Nachin, L., Diez, A. & Nyström, T. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6, 140–145 (2003).
Kerk, D., Bulgrien, J., Smith, D. W. & Gribskov, M. Arabidopsis proteins containing similarity to the universal stress protein domain of bacteria. Plant Physiol. 131, 1209–1219 (2003).
van Dongen, J. T., Schurr, U., Pfister, M., & Geigenberger, P. Phloem metabolism and function have to cope with low internal oxygen. Plant Physiol. 131, 1529–1543 (2003).
Nachin, L., Brive, L., Persson, K. C., Svensson, P. & Nyström, T. Heterodimer formation within universal stress protein classes revealed by an in silico and experimental approach. J. Mol. Biol. 380, 340–450 (2008).
Foreman, J. et al.. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446 (2003).
Li, H., Shen, J. J., Zheng, Z. L., Lin, Y. & Yang, Z. The RopGTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol. 126, 670–684 (2001).
Jones, M. A., Raymond, M. J., & Smirnoff, N. Analysis of the root hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root hair development in Arabidopsis. Plant J. 4, 83–100 (2006).
Jones, M. A. The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14, 763–776 (2002).
Nibau, C., Wu, H. & Cheung, A. Y. Rac/RopGTPases: ‘hubs’ for signal integration and diversification in plants. Trends Plant Sci. 11, 309–315 (2006).
Yang, Z. & Ying, F. ROP/RAC GTPase signaling. Curr. Opin. Plant Biol. 10, 490–494 (2007).
Sagi, M. & Fluhr, R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141, 336–340 (2006).
Wong, H. L. et al. Regulation of rice NADPH oxidase by binding of RacGTPase to its N-terminal extension. Plant Cell 19, 4022–4034 (2007).
Ueoka-Nakanishi, H. et al. Thioredoxin h regulates calcium dependent protein kinases in plasma membranes. FEBS J. 280, 3220–3231 (2013).
Zheng, Z. L. & Yang, Z. The RopGTPase: an emerging signaling switch in plants. Plant Mol. Biol. 44, 1–9 (2000).
Marmagne, A. et al. A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Mol. Cell. Prot. 6, 1980–1996 (2007).
Mitra, S. K., Gantt, J. A., Ruby, J. F., Clouse, S. D. & Goshe, M. B. Membrane proteomic analysis of Arabidopsis thaliana using alternative solubilization techniques. J. Prot. Res. 6, 1933–1950 (2007).
Di Rienzo, C., Piazza, V., Gratton, E., Beltram, F. & Cardarelli, F. Probing short-range protein Brownian motion in the cytoplasm of living cells. Nature Comm. 5, 5891 (2014).
Hao, H. et al. Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell 26, 1729–1745 (2014).
Keller, T. et al. A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell 10, 255–266 (1998).
Boes, N., Schreiber, K., Hartig, E., Jaensch, L. & Schobert, M. The Pseudomonas aeruginosa universal stress protein PA4352 is essential for surviving anaerobic energy stress. J. Bacteriol. 188, 6529–6538 (2006).
O'Toole, R. & Williams, H. D. Universal stress proteins and Mycobacterium tuberculosis. Res. Microbiol. 154, 387–392 (2003).
Loukehaich, R. et al. SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J Exp. Bot. 63, 5593–5606 (2012).
Maqbool, A., Zahur, M., Husnain, T. & Riazuddin, S. GUSP1 and GUSP2, two drought-responsive genes in Gossypium arboreum have homology to universal stress proteins. Plant Mol. Biol. Rep. 27, 109–114 (2009).
Sauter, M., Rzewuski, G., Marwedel, T. & Lorbiecke, R. The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 53, 2325–2331 (2002).
Subbaiah, C. C., Bush, D. S. & Sachs, M. M. Elevation of cytosolic calcium precedes anoxic gene expression in maize suspension-cultured cells. Plant Cell 6, 1747–1762 (1994).
Subbaiah, C. C., Zhang, J. & Sachs, M. M. Involvement of intracellular calcium in anaerobic gene expression and survival of maize seedlings. Plant Physiol. 105, 369–376 (1994).
Sedbrook, J. C., Kronebusch, P. J., Borisy, G. G., Trewavas, A. J. & Masson, P. H. Transgenic AEQUORIN reveals organ-specific cytosolic Ca2+ responses to anoxia in Arabidopsis thaliana seedlings. Plant Physiol. 111, 243–257 (1996).
Kobayashi, M. et al. Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19, 1065–1080 (2007).
Hann, L. W. et al. Regulation of rice NADPH oxidase by binding of RacGTPase to its N-terminal extension. Plant Cell 19, 4022–4034 (2007).
van Dongen, J. T. & Licausi, F. Oxygen sensing and signaling. Annu. Rev. Plant Biol. 66, 345–367 (2015).
Mithran, M., Paparelli, E., Novi, G., Perata, P. & Loreti, E. Analysis of the role of the pyruvate decarboxylase gene family in Arabidopsis thaliana under low-oxygen conditions. Plant Biol. 16, 28–23 (2014).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protoc. 215, 65–72 (2007).
Perata, P., Matsukura, C., Vernieri, P. & Yamaguchi, J. Sugar repression of a gibberellin-dependent signaling pathway in barley embryos. Plant Cell 9, 2197–2208 (1997).
Arvidsson, S., Kwasniewski, M., Riano-Pachon, D. M. & Mueller-Roeber, B. QuantPrime – a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 9, 465 (2008).
Loreti, E., Alpi, A. & Perata, P. Glucose and disaccharide-sensing mechanisms modulate the expression of alpha-amylase in barley embryos. Plant Physiol. 123, 939–948 (2000).
Nakagami, H., Soukupová, H., Schikora, A., Zárský, V. & Hirt, H. A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol. Chem. 281, 38697–38704 (2006).
Acknowledgements
We thank J. Bailey-Serres for reviewing an early version of this manuscript, A. Galli for helpful suggestions about the Y2H assay and C. Kiferle for helpful assistance in data analysis. We acknowledge Z. Yang (University of California, Riverside) for providing us with the plasmid carrying the coding sequence for the RIC1-Maltose Binding Protein (MBP) fusion protein.
Author information
Authors and Affiliations
Contributions
S.G., E.L., L.B., S.P., C.P. and F.L. performed the molecular biology experiments, Y2H, BiFC and split-luciferase assays. G.N. and S.P. obtained the mutants and performed the phenotypic characterization of the mutants and transgenic lines. G.N. performed the submergence and hypoxia tolerance assays. E.L. and P.P. performed the transient expression analysis in Nicotiana. V.B. and C.P. performed the ROP pull-down assays. F.C. designed, performed and analysed the FRAP experiments. P.P., S.G. and E.L. designed the experiments. P.P. and E.L. wrote the manuscript. All authors discussed and commented on the content of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Gonzali, S., Loreti, E., Cardarelli, F. et al. Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nature Plants 1, 15151 (2015). https://doi.org/10.1038/nplants.2015.151
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2015.151
This article is cited by
-
Deepening the knowledge of universal stress proteins in Haloferax mediterranei
Applied Microbiology and Biotechnology (2024)
-
Identification of universal stress proteins in wheat and functional characterization during abiotic stress
Plant Cell Reports (2023)
-
Genome-wide identification and expression profiling of genes encoding universal stress proteins (USP) identify multi-stress responsive USP genes in Arabidopsis thaliana
Plant Physiology Reports (2019)
-
Both AtrbohD and AtrbohF are essential for mediating responses to oxygen deficiency in Arabidopsis
Plant Cell Reports (2017)