Abstract
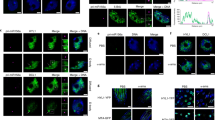
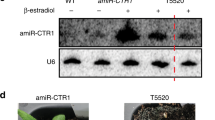
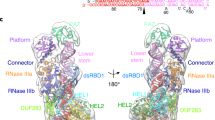
MicroRNAs (miRNAs) are small regulatory RNAs produced by Dicer proteins that regulate gene expression in development and adaptive responses to the environment1–4. In animals, the degree of base pairing between a miRNA and its target messenger RNA seems to determine whether the regulation occurs through cleavage or translation inhibition1. In contrast, the selection of regulatory mechanisms is independent of the degree of mismatch between a plant miRNA and its target transcript5. However, the components and mechanism(s) that determine whether a plant miRNA ultimately regulates its targets by guiding cleavage or translational inhibition are unknown6. Here we show that the form of regulatory action directed by a plant miRNA is determined by DRB2, a DICER-LIKE1 (DCL1) partnering protein. The dependence of DCL1 on DRB1 for miRNA biogenesis is well characterized7–9, but we show that it is only required for miRNA-guided transcript cleavage. We found that DRB2 determines miRNA-guided translational inhibition and represses DRB1 expression, thereby allowing the active selection of miRNA regulatory action. Furthermore, our results reveal that the core silencing proteins ARGONAUTE1 (AGO1) and SERRATE (SE) are highly regulated by miRNA-guided translational inhibition. DRB2 has been remarkably conserved throughout plant evolution, raising the possibility that translational repression is the ancient form of miRNA-directed gene regulation in plants, and that Dicer partnering proteins, such as human TRBP, might play a similar role in other eukaryotic systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ameres, S. L. & Zamore, P. D. Diversifying microRNA sequence and function. Nature Rev. Mol. Cell Biol. 14, 475–488 (2013).
Bologna, N. G. & Voinnet, O. Diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65, 473–503 (2014).
Sunkar, R., Li, Y-F. & Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 17, 196–203 (2012).
Poethig, R. S. The past, present, and future of vegetative phase change. Plant Physiol. 154, 541–544 (2010).
Brodersen, P. et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190 (2008).
Mallory, A. & Vaucheret, H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22, 3879–3889 (2010).
Han, M-H., Goud, S., Song, L. & Fedoroff, N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl Acad. Sci. USA 101, 1093–1098 (2004).
Hiraguri, A. et al. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 57, 173–188 (2005).
Eamens, A. L., Smith, N. A., Curtin, S. J., Wang, M-B. & Waterhouse, P. M. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15, 2219–2235 (2009).
Eamens, A. L., Kim, K. W., Curtin, S. J. & Waterhouse, P. M. DRB2 Is Required for MicroRNA Biogenesis in Arabidopsis thaliana. PLoS ONE 7, e35933 (2012).
Curtin, S. J. et al. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 582, 2753–2760 (2008).
Dugas, D. & Bartel, B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol. Biol. 67, 403–417 (2008).
Lanet, E. et al. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21, 1762–1768 (2009).
Waterhouse, P. M., Wang, M-B. & Lough, T. Gene silencing as an adaptive defence against viruses. Nature 411, 834–842 (2001).
Rajagopalan, R., Vaucheret, H., Trejo, J. & Bartel, D. P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425 (2006).
Raja, P., Jackel, J. N., Li, S., Heard, I. M. & Bisaro, D. M. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. JVI, 02305–02313 (2013).
Chen, X. A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development. Science 303, 2022–2025 (2004).
Yang, L., Wu, G. & Poethig, R. S. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc. Natl Acad. Sci. USA 109, 315–320 (2012).
Xie, Z., Kasschau, K. D. & Carrington, J. C. Negative feedback regulation of Dicer-like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13, 784–789 (2003).
Eamens, A. L., Wook Kim, K. & Waterhouse, P. M. DRB2, DRB3 and DRB5 function in a non-canonical microRNA pathway in Arabidopsis thaliana. Plant Signal. Behav. 7, 1224–1229 (2012).
Floyd, S. K. & Bowman, J. L. Gene regulation: ancient microRNA target sequences in plants. Nature 428, 485–486 (2004).
Horman, S. R. et al. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol. Cell 50, 356–367 (2013).
Bouquin, T., Mattsson, O., Næsted, H., Foster, R. & Mundy, J. The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 116, 791–801 (2003).
Martin, T., Oswald, O. & Graham, I. A. Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 128, 472–481 (2002).
Coutu, C., Brandle, J., Brown, D. & Brown, K. pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res. 16, 771–781 (2007).
Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987).
Stocks, M. B. et al. The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28, 2059–2061 (2012).
Skirycz, A. et al. A Reciprocal 15N-labeling proteomic analysis of expanding Arabidopsis leaves subjected to osmotic stress indicates importance of mitochondria in preserving plastid functions. J. Proteome Res. 10, 1018–1029 (2010).
Arsova, B., Zauber, H. & Schulze, W. X. Precision, proteome coverage, and dynamic range of arabidopsis proteome profiling using 15N metabolic labeling and label-free approaches. Mol. Cell. Proteomics 11, 619–628 (2012).
Schaff, J. E., Mbeunkui, F., Blackburn, K., Bird, D. M. & Goshe, M. B. SILIP: a novel stable isotope labeling method for in planta quantitative proteomic analysis. Plant J. 56, 840–854 (2008).
Acknowledgements
We thank K. Nakasugi for deep sequencing data analysis and D. Barton for GUS imaging, A. Tay for statistical analysis of proteomics data, A.V.R. Ribeiro for graphical assistance and T. Roberts and R. Hellens for critical reading of our manuscript. We acknowledge M. Raftery, L. Zhong and S. Liu Lau for their maintenance of the orbitrap mass spectrometers at the UNSW Bioanalytical Mass Spectrometry Facility. PMW acknowledges support as a Federation Fellow from the Australian Research Council and contributions from the University of Sydney, CSIRO and QUT. M.R.W. acknowledges support from the Australian Research Council, the Australian Federal Government EIF Super Science Scheme and the University of New South Wales. G.H-S. was the recipient of an Australian Research Council Australian Postdoctoral Research Fellowship and funds from the University of New South Wales ECR Scheme. R.S.R. was the recipient of an Australian Postgraduate Award.
Author information
Authors and Affiliations
Contributions
G.H-S., R.S.R. and M.R.W. conceived and designed the proteomics experiments; G.H-S. performed the mass spectrometry experiments and data analysis; R.S.R., A.L.E. and P.M.W. conceived all other experiments; A.L.E. performed miR863 northern blots; R.S.R. designed and performed all other experiments; R.S.R. and P.M.W. interpreted the data; R.S.R., A.L.E. and P.M.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Reis, R., Hart-Smith, G., Eamens, A. et al. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nature Plants 1, 14027 (2015). https://doi.org/10.1038/nplants.2014.27
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2014.27
This article is cited by
-
Plant gene silencing signals move from the phloem to influence gene expression in shoot apical meristems
BMC Plant Biology (2022)
-
Identification and profiling of narrow-leafed lupin (Lupinus angustifolius) microRNAs during seed development
BMC Genomics (2019)
-
Mechanisms of microRNA-mediated gene regulation in unicellular model alga Chlamydomonas reinhardtii
Biotechnology for Biofuels (2018)
-
Biological significance, computational analysis, and applications of plant microRNAs
Acta Physiologiae Plantarum (2018)
-
The evolutionary origin of plant and animal microRNAs
Nature Ecology & Evolution (2017)