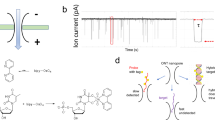
Abstract
MicroRNAs are short RNA molecules that regulate gene expression, and have been investigated as potential biomarkers because their expression levels are correlated with various diseases. However, detecting microRNAs in the bloodstream remains difficult because current methods are not sufficiently selective or sensitive. Here, we show that a nanopore sensor based on the α-haemolysin protein can selectively detect microRNAs at the single molecular level in plasma samples from lung cancer patients without the need for labels or amplification of the microRNA. The sensor, which uses a programmable oligonucleotide probe to generate a target-specific signature signal, can quantify subpicomolar levels of cancer-associated microRNAs and can distinguish single-nucleotide differences between microRNA family members. This approach is potentially useful for quantitative microRNA detection, the discovery of disease markers and non-invasive early diagnosis of cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Carthew, R. W. & Sontheimer, E. J. Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009).
Inui, M., Martello, G. & Piccolo, S. MicroRNA control of signal transduction. Nature Rev. Mol. Cell Biol. 11, 252–263 (2010).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Kozomara, A. & Griffiths-Jones, S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 (2011).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Diederichs, S. & Haber, D. A. Dual role for argonautes in MicroRNA processing and posttranscriptional regulation of MicroRNA expression. Cell 131, 1097–1108 (2007).
Garzon, R., Calin, G. A. & Croce, C. M. MicroRNAs in cancer. Ann. Rev. Med. 60, 167–179 (2009).
Ortholan, C. et al. MicroRNAs and lung cancer: new oncogenes and tumor suppressors, new prognostic factors and potential therapeutic targets. Curr. Med. Chem. 16, 1047–1061 (2009).
Calin, G. A. & Croce, C. M. MicroRNA signatures in human cancers. Nature Rev. Cancer 6, 857–866 (2006).
Mitchell, P. S. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA 105, 10513–10518 (2008).
Kosaka, N. et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452 (2010).
Rabinowits, G., Gerçel-Taylor, C., Day, J. M., Taylor, D. D. & Kloecker, G. H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer 10, 42–46 (2009).
Rosell, R., Wei, J. & Taron, M. Circulating MicroRNA signatures of tumor-derived exosomes for early diagnosis of non-small-cell lung cancer. Clin. Lung Cancer 10, 8–9 (2009).
Chen, C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 (2005).
Li, W. & Ruan, K. MicroRNA detection by microarray. Anal. Bioanal. Chem. 394, 1117–1124 (2009).
Hunt, E. A., Goulding, A. M. & Deo, S. K. Direct detection and quantification of microRNAs. Anal. Biochem. 387, 1–12 (2009).
Yendamuri, S. & Kratzke, R. MicroRNA biomarkers in lung cancer: MiRacle or quagMiRe? Transl. Res. 157, 209–215 (2011).
Neely, L. A. et al. A single-molecule method for the quantitation of microRNA gene expression. Nature Methods 3, 41–46 (2006).
Bayley, H. & Jayasinghe, L. Functional engineered channels and pores — (Review). Mol. Membrane Biol. 21, 209–220 (2004).
Gu, L. Q. & Shim, J. W. Single molecule sensing by nanopores and nanopore devices. Analyst 135, 441–451 (2010).
Howorka, S. & Siwy, Z. Nanopore analytics: sensing of single molecules. Chem. Soc. Rev. 38, 2360–2384 (2009).
Ma, L. & Cockroft, S. L. Biological nanopores for single-molecule biophysics. Chembiochem 11, 25–34 (2010).
Movileanu, L. Interrogating single proteins through nanopores: challenges and opportunities. Trends Biotechnol. 27, 333–341 (2009).
Olasagasti, F. et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nature Nanotech. 5, 798–806 (2010).
Bayley, H. Sequencing single molecules of DNA. Current Opin. Chem. Biol. 10, 628–637 (2006).
Branton, D. et al. The potential and challenges of nanopore sequencing. Nature Biotechnol. 26, 1146–1153 (2008).
Wanunu, M. et al. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Natur Nanotech. 5, 807–814 (2010).
Song, L. Z. et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866 (1996).
Kasianowicz, J. J., Brandin, E., Branton, D. & Deamer, D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl Acad. Sci. USA 93, 13770–13773 (1996).
Meller, A., Nivon, L. & Branton, D. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 86, 3435–3438 (2001).
Mitchell, N. & Howorka, S. Chemical tags facilitate the sensing of individual DNA strands with nanopores. Angew. Chem. Int. Ed. 47, 5565–5568 (2008).
Meller, A., Nivon, L., Brandin, E., Golovchenko, J. & Branton, D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl Acad. Sci. USA 97, 1079–1084 (2000).
Donnem, T. et al. Prognostic impact of miR-155 in non-small cell lung cancer evaluated by in situ hybridization. J. Transl. Med. 9, 6 (2011).
Mathé, J., Visram, H., Viasnoff, V., Rabin, Y. & Meller, A. Nanopore unzipping of individual DNA hairpin molecules. Biophys. J. 87, 3205–3212 (2004).
Sauer-Budge, A. F., Nyamwanda, J. A., Lubensky, D. K. & Branton, D. Unzipping kinetics of double-stranded DNA in a nanopore. Phys. Rev. Lett. 90, 238101 (2003).
Butler, T. Z., Gundlach, J. H. & Troll, M. Ionic current blockades from DNA and RNA molecules in the alpha-hemolysin nanopore. Biophys. J. 93, 3229–3240 (2007).
Mathé, J., Aksimentiev, A., Nelson, D. R., Schulten, K. & Meller, A. Orientation discrimination of single-stranded DNA inside the alpha-hemolysin membrane channel. Proc. Natl Acad. Sci. USA 102, 12377–12382 (2005).
Purnell, R. F., Mehta, K. K. & Schmidt, J. J. Nucleotide identification and orientation discrimination of DNA homopolymers immobilized in a protein nanopore. Nano. Lett. 8, 3029–3034 (2008).
Wanunu, M., Morrison, W., Rabin, Y., Grosberg, A. Y. & Meller, A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nature Nanotech. 5, 160–165 (2010).
Maglia, G., Restrepo, M. R., Mikhailova, E. & Bayley, H. Enhanced translocation of single DNA molecules through α-hemolysin nanopores by manipulation of internal charge. Proc. Natl. Acad. Sci. USA 105, 19720–19725 (2008).
Cho, W. C. Role of miRNAs in lung cancer. Expert Rev. Mol. Diagn. 9, 773–776 (2009).
Howorka, S., Cheley, S. & Bayley, H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nature Biotechnol. 19, 636–639 (2001).
Nakane, J., Wiggin, M. & Marziali, A. A nanosensor for transmembrane capture and identification of single nucleic acid molecules. Biophys. J. 87, 615–621 (2004).
Vercoutere, W. et al. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nature Biotechnol. 19, 248–252 (2001).
Landi, M. T. et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin. Cancer Res. 16, 430–441 (2010).
Silvestri, G. A., Alberg, A. J. & Ravenel, J. The changing epidemiology of lung cancer with a focus on screening. Br. Med. J. 339, 63053 (2009).
Patnaik, S. K., Kannisto, E., Knudsen, S. & Yendamuri, S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 70, 36–45 (2010).
Yanaihara, N. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198 (2006).
Singer, A. et al. Nanopore based sequence specific detection of duplex DNA for genomic profiling. Nano Lett. 10, 738–742 (2010).
Bayley, H. et al. Droplet interface bilayers. Mol. Biosyst. 4, 1191–1208 (2008).
Acknowledgements
The authors thank K. Gillis, T-C. Hwang, S-J. Chen, F. Hsieh and M. Milanick for invaluable discussions on experimental design and data analysis. This investigation was partially supported by grants from the National Science Foundation 0546165 (L-Q.G.), the National Institutes of Health GM079613 (L-Q.G.) and the University of Missouri Intellectual Property Fast Track Initiative (A8881, M.X.W.), and was conducted in a facility that was constructed with support from the Research Facilities Improvement Program grant no. C06-RR-016489-01 from the National Centre for Research Resources, National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Y.W. designed and performed the nanopore experiments, collected and analysed the nanopore data, and co-wrote the manuscript. D.Z. designed and performed the qRT-PCR experiments, analysed the qRT-PCR data and co-wrote the manuscript. Q.T. performed molecular biology experiments, including protein synthesis. M.X.W. conceived the qRT-PCR experiments, provided the patients' samples and co-wrote the manuscript. L-Q.G. conceived the principal idea, designed the nanopore experiments and wrote the manuscript. All the authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 691 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Zheng, D., Tan, Q. et al. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nature Nanotech 6, 668–674 (2011). https://doi.org/10.1038/nnano.2011.147
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2011.147
This article is cited by
-
Single cell quantification of microRNA from small numbers of non-invasively sampled primary human cells
Communications Biology (2023)
-
PD-L1-driven efficient enrichment and elimination of circulating cancer cells by magnetic MoSe2 nanosheet
Nano Research (2023)
-
A revised analysis of current and emerging Nano suspension technological approaches for cardiovascular medicine
Beni-Suef University Journal of Basic and Applied Sciences (2022)
-
A reversibly gated protein-transporting membrane channel made of DNA
Nature Communications (2022)
-
A click chemistry amplified nanopore assay for ultrasensitive quantification of HIV-1 p24 antigen in clinical samples
Nature Communications (2022)