Abstract
To achieve accurate spatiotemporal patterns of gene expression, RNA-binding proteins (RBPs) guide nuclear processing, intracellular trafficking and local translation of target mRNAs. In neurons, RBPs direct transport of target mRNAs to sites of translation in remote axons and dendrites. However, it is not known whether an individual RBP coordinately regulates multiple mRNAs within these morphologically complex cells. Here we identify SFPQ (splicing factor, poly-glutamine rich) as an RBP that binds and regulates multiple mRNAs in dorsal root ganglion sensory neurons and thereby promotes neurotrophin-dependent axonal viability. SFPQ acts in nuclei, cytoplasm and axons to regulate functionally related mRNAs essential for axon survival. Notably, SFPQ is required for coassembly of LaminB2 (Lmnb2) and Bclw (Bcl2l2) mRNAs in RNA granules and for axonal trafficking of these mRNAs. Together these data demonstrate that SFPQ orchestrates spatial gene expression of a newly identified RNA regulon essential for axonal viability.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Martin, K.C. & Ephrussi, A. mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730 (2009).
Mitchell, S.F. & Parker, R. Principles and properties of eukaryotic mRNPs. Mol. Cell 54, 547–558 (2014).
Blackinton, J.G. & Keene, J.D. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin. Cell Dev. Biol. 34, 44–54 (2014).
Keene, J.D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533–543 (2007).
Holt, C.E. & Schuman, E.M. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80, 648–657 (2013).
Jung, H., Gkogkas, C.G., Sonenberg, N. & Holt, C.E. Remote control of gene function by local translation. Cell 157, 26–40 (2014).
Yarosh, C.A., Iacona, J.R., Lutz, C.S. & Lynch, K.W. PSF: nuclear busy-body or nuclear facilitator? Wiley Interdiscip. Rev. RNA 6, 351–367 (2015).
Dong, X., Sweet, J., Challis, J.R., Brown, T. & Lye, S.J. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol. Cell. Biol. 27, 4863–4875 (2007).
Hirose, T. et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 25, 169–183 (2014).
Patton, J.G., Porro, E.B., Galceran, J., Tempst, P. & Nadal-Ginard, B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7, 393–406 (1993).
Danckwardt, S. et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 26, 2658–2669 (2007).
Hall-Pogar, T., Liang, S., Hague, L.K. & Lutz, C.S. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA 13, 1103–1115 (2007).
Chanas-Sacré, G. et al. Identification of PSF, the polypyrimidine tract-binding protein-associated splicing factor, as a developmentally regulated neuronal protein. J. Neurosci. Res. 57, 62–73 (1999).
Lowery, L.A., Rubin, J. & Sive, H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev. Dyn. 236, 1347–1357 (2007).
Kanai, Y., Dohmae, N. & Hirokawa, N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 (2004).
Kunde, S.A. et al. The X-chromosome-linked intellectual disability protein PQBP1 is a component of neuronal RNA granules and regulates the appearance of stress granules. Hum. Mol. Genet. 20, 4916–4931 (2011).
Ray, D. et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 (2013).
Deglincerti, A. & Jaffrey, S.R. Insights into the roles of local translation from the axonal transcriptome. Open Biol. 2, 120079 (2012).
Yoon, B.C. et al. Local translation of extranuclear lamin B promotes axon maintenance. Cell 148, 752–764 (2012).
Cosker, K.E., Pazyra-Murphy, M.F., Fenstermacher, S.J. & Segal, R.A. Target-derived neurotrophins coordinate transcription and transport of bclw to prevent axonal degeneration. J. Neurosci. 33, 5195–5207 (2013).
Andreassi, C. et al. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat. Neurosci. 13, 291–301 (2010).
Cox, L.J., Hengst, U., Gurskaya, N.G., Lukyanov, K.A. & Jaffrey, S.R. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat. Cell Biol. 10, 149–159 (2008).
Snider, W.D. Functions of the neurotrophins during nervous system development: what the knockouts are teaching us. Cell 77, 627–638 (1994).
Zhang, H.L., Singer, R.H. & Bassell, G.J. Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J. Cell Biol. 147, 59–70 (1999).
Courchesne, S.L., Karch, C., Pazyra-Murphy, M.F. & Segal, R.A. Sensory neuropathy attributable to loss of Bcl-w. J. Neurosci. 31, 1624–1634 (2011).
Batish, M., van den Bogaard, P., Kramer, F.R. & Tyagi, S. Neuronal mRNAs travel singly into dendrites. Proc. Natl. Acad. Sci. USA 109, 4645–4650 (2012).
De Rubeis, S. & Bagni, C. Fragile X mental retardation protein control of neuronal mRNA metabolism: insights into mRNA stability. Mol. Cell. Neurosci. 43, 43–50 (2010).
Okano, H. et al. Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 306, 349–356 (2005).
Gagnon, J.A. & Mowry, K.L. Molecular motors: directing traffic during RNA localization. Crit. Rev. Biochem. Mol. Biol. 46, 229–239 (2011).
Hirokawa, N. & Takemura, R. Molecular motors and mechanisms of directional transport in neurons. Nat. Rev. Neurosci. 6, 201–214 (2005).
Spillane, M., Ketschek, A., Merianda, T.T., Twiss, J.L. & Gallo, G. Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep. 5, 1564–1575 (2013).
Campenot, R.B. Development of sympathetic neurons in compartmentalized cultures. II. Local control of neurite survival by nerve growth factor. Dev. Biol. 93, 13–21 (1982).
Gerber, A.P., Herschlag, D. & Brown, P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2, E79 (2004).
García-Rodríguez, L.J., Gay, A.C. & Pon, L.A. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J. Cell Biol. 176, 197–207 (2007).
Ule, J. et al. CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215 (2003).
Farris, S., Lewandowski, G., Cox, C.D. & Steward, O. Selective localization of arc mRNA in dendrites involves activity- and translation-dependent mRNA degradation. J. Neurosci. 34, 4481–4493 (2014).
Lee, M. et al. The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 43, 3826–3840 (2015).
Hüttelmaier, S. et al. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512–515 (2005).
Buxadé, M., Morrice, N., Krebs, D.L. & Proud, C.G. The PSF.p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor alpha. J. Biol. Chem. 283, 57–65 (2008).
Ke, Y.D. et al. Tau-mediated nuclear depletion and cytoplasmic accumulation of SFPQ in Alzheimer's and Pick's disease. PLoS One 7, e35678 (2012).
Tapia-Páez, I., Tammimies, K., Massinen, S., Roy, A.L. & Kere, J. The complex of TFII-I, PARP1, and SFPQ proteins regulates the DYX1C1 gene implicated in neuronal migration and dyslexia. FASEB J. 22, 3001–3009 (2008).
Kubota, M. et al. Therapeutic implications of down-regulation of cyclophilin D in bipolar disorder. Int. J. Neuropsychopharmacol. 13, 1355–1368 (2010).
Hempstead, B.L. et al. Overexpression of the trk tyrosine kinase rapidly accelerates nerve growth factor-induced differentiation. Neuron 9, 883–896 (1992).
Ross, A.J. et al. Testicular degeneration in Bclw-deficient mice. Nat. Genet. 18, 251–256 (1998).
Pazyra-Murphy, M.F. & Segal, R.A. Preparation and maintenance of dorsal root ganglia neurons in compartmented cultures. J. Vis. Exp. 10.3791/951 (2008).
Fenstermacher, S.J., Pazyra-Murphy, M.F. & Segal, R.A. Campenot cultures and microfluidics provide complementary platforms for spatial study of dorsal root ganglia neurons. Springer Protocols: Neuromethods 103, 105–124 (2015).
Watson, F.L. et al. Rapid nuclear responses to target-derived neurotrophins require retrograde transport of ligand-receptor complex. J. Neurosci. 19, 7889–7900 (1999).
Aguet, F., Antonescu, C.N., Mettlen, M., Schmid, S.L. & Danuser, G. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell 26, 279–291 (2013).
Lachmanovich, E. et al. Co-localization analysis of complex formation among membrane proteins by computerized fluorescence microscopy: application to immunofluorescence co-patching studies. J. Microsc. 212, 122–131 (2003).
Mendoza, M.C. et al. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol. Cell 41, 661–671 (2011).
Acknowledgements
We thank members of the Segal laboratory, A. Eisner, M. McClintock and M. Greenberg for discussions, and G. McGregor (University of California, Irvine) for providing Bcl2l2−/− mice. This work was supported by US National Institutes of Health grants R01 NS050674 and R01 MH091662 to R.A.S. and F31 NS077620 to S.J.F.; Harvard/MIT Joint Research Program in Basic Neuroscience Grant to R.A.S. and M. Heiman; fellowships from the Harvard Mahoney, the Harvard NeuroDiscovery Center and the Alice and Joseph E. Brooks fund to K.E.C.; and a fellowship from the Victoria Quan fund to S.J.F.
Author information
Authors and Affiliations
Contributions
K.E.C., S.J.F. and R.A.S. designed experiments and wrote the manuscript. K.E.C., S.J.F. and M.F.P.-M. performed compartmentalized culture experiments. K.E.C. performed formaldehyde cross-linking experiments. M.F.P.-M. performed axonal degeneration and protein transfection experiments. H.L.E. analyzed FISH data. K.E.C. and S.J.F. performed all other experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 SFPQ is in axons and binds axonal mRNAs
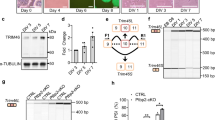
(a) Previously identified axonal mRNAs analyzed for SFPQ-binding motifs10, 20. (b) qRT-PCR analysis of axonal genes expressed in Trk-PC12 cells and DRG sensory neurons normalized to gapdh mRNA. Data shows mean ± SEM, n = 3 experiments; *P < 0.05 (Unpaired two-tailed t-test; P = 0.3, t(3) = 1.25 for laminb2, P = 0.54, t(3) = 0.68 for bclw, P = 0.028, t(3) = 3.9 for impa1, P = 0.26, t(3) = 1.37 for creb, P = 0.33, t(3) =1.17 for β-actin, P = 0.31, t(3) = 1.22 for cox4, P = 0.01, t(3) = 5.62 for smad, P = 0.06, t(3) = 3.01 for rpl4). (c) qRT-PCR analysis of SFPQ-precipitated mRNAs from Trk-PC12 cells in the absence or presence of NGF stimulation following formaldehyde crosslinking. Data, normalized to no antibody control, shows mean ± SEM, n = 4 experiments; *P < 0.05 (Paired two-tailed t-test; P = 0.05, t(3) = 2.44 for laminb2, P = 0.04, t(3) = 2.48 for bclw, P = 0.01, t(3) = 4.03 for impa1, P = 0.05, t(3) = 2.53 for creb, P = 0.35, t(3) = 0.42 for β-actin, P = 0.08, t(3) = 1.85 for cox4, P = 0.4, t(2) = 0.29 for smad, P = 0.2, t(2) = 1.26 for rpl4, P = 0.3, t(3) = 0.6 for gapdh). (d) SFPQ immunostaining of whole-mount DRG at P0 shows SFPQ within neuronal nuclei (white arrow heads). Scale bar, 100 µm. (e) Western blot of sensory neurons cultured in compartmented cultures and lysates from cell bodies (CB) and distal axons (DA) probed for SFPQ, GAPDH and Histone with SFPQ protein quantification; data shows mean ± SEM, n = 3 experiments. (f) SFPQ immunostaining of cultured sensory neurons expressing control or SFPQ shRNA. Scale bar, 10 µm. (g) SFPQ immunostaining of cultured sensory neuron axons expressing control or SFPQ shRNA. Scale bar, 5 µm.
Supplementary Figure 2 Validation of single-molecule FISH probes
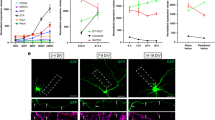
(a) qRT-PCR analysis of sfpq mRNA, normalized to gapdh, in cell bodies of sensory neurons grown in compartmented cultures expressing control or SFPQ shRNA. Data, normalized to control, shows mean ± SEM, n = 8; *P = 0.0003, t(7) = 6.74 (Unpaired two-tailed t-test). (b) Single-molecule FISH for laminb2 mRNA in cells expressing control or laminb2 shRNA (max projection). Quantification of laminb2 mRNA spots per cell of a single image stack; data shows mean ± SEM, n = 458 for shCntrl and n = 638 for shLmnB2 cells from 3 experiments; *P < 0.0001, t(20) = 5.19 (Unpaired two-tailed t-test). Scale bar, 10 μm. (c) Single-molecule FISH for bclw mRNA on frozen sections of DRG (L4) from bclw+/+ and bclw−/− mice (6 months), stained for Tuj1 and DAPI. Scale bar, 100 μm. (d) Number of single-molecule FISH mRNA puncta per μm2 in DA following 2h neurotrophin stimulation of DA. Data shows mean ± SEM, n = 3 individual microfluidic cultures; *P = 0.11, t(79) = 1.58 (Unpaired two-tailed t-test between Cn and NT). (e) Number of single-molecule FISH mRNA puncta per μm2 in DA of sensory neurons expressing SFPQ shRNA. Data shows mean ± SEM, n = 3 individual microfluidic cultures; P = 0.422, t(68) = 0.82 (Unpaired two-tailed t-test between shCntrl and shSFPQ).
Supplementary Figure 3 Reverse colocalization analysis
(a,b) Reverse colocalization analysis from Fig. 4c,d in control (a) and shSFPQ (b) conditions. Normalized density is frequency of adjacent mRNAs at each distance on x-axis normalized to the alternate frequency expected by chance (reverse randomization).
Supplementary Figure 4 The Bcl2l2 3′ UTR is critical for neurotrophin-dependent axonal localization
(a) qRT-PCR analysis of egfp mRNA levels in sensory neurons grown in compartmented cultures expressing eGFP or eGFP-Bclw-3’UTR following 2h neurotrophin stimulation of distal axons (DA); data shows mean ± SEM, n = 6 experiments; P = 0.05, t(10) = 2.18 (Unpaired two-tailed t-test). In DA, neurotrophin-dependent regulation of egfp mRNA occurs only in the presence of the bclw 3’UTR.
Supplementary Figure 5 SFPQ and cell body apoptosis
(a) DAPI staining of cell bodies (CB) of sensory neurons grown in compartmented cultures expressing control or SFPQ shRNA and quantification of percentage condensed nuclei (arrow heads). Scale bar, 40 μm. n = 4746 cells for shCntrl and n = 3817 cells for shSFPQ from 3 experiments; n.s., not significant (two-way ANOVA with Bonferroni correction; P = 0.49, F(1,80) = 0.48). (b) Western blot of LaminB2 in CB of sensory neurons grown in compartmented cultures expressing control or laminb2 shRNA. Quantification of protein levels normalized to GAPDH, data shows mean ± SEM, n = 3 experiments; *P < 0.05 (Unpaired two-tailed t-test; P = 0.003, t(4) = 6.36). (c) Western blot for His after selective introduction of His-tagged Bclw protein into distal axons (DA) of sensory neurons grown in compartmented cultures.
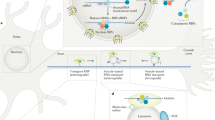
Supplementary Figure 6 Model for SFPQ and the axonal mRNA regulon
SFPQ binds both bclw mRNA and laminb2 mRNA enabling neurotrophin-dependent localization to distal axons. Following neurotrophin stimulation of distal axons, SFPQ binds bclw mRNA enabling nuclear export. In the cytoplasm, SFPQ and bclw join with SFPQ-bound laminb2 and/or other axonally targeted mRNAs to form an RNA transport granule that traffics to axons. There, mRNAs are locally translated at mitochondria to generate Bclw and LaminB2 protein that promote axon survival.
Supplementary Figure 7 Full-length blots and gels
(a) Figure 2e. (b) Figure 3a. (c) Figure 5a. (d) Figure 5b. (e) Figure 5c. (f) Figure 5d. (g) Figure 6a. (h) Supp. Figure 1e. (i) Supp. Figure 5b. (j) Supp. Figure 5c.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Table 1 (PDF 1883 kb)
Rights and permissions
About this article
Cite this article
Cosker, K., Fenstermacher, S., Pazyra-Murphy, M. et al. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat Neurosci 19, 690–696 (2016). https://doi.org/10.1038/nn.4280
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4280
This article is cited by
-
Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy
Journal of Human Genetics (2023)
-
SFPQ rescues F508del-CFTR expression and function in cystic fibrosis bronchial epithelial cells
Scientific Reports (2021)
-
The functional organization of axonal mRNA transport and translation
Nature Reviews Neuroscience (2021)
-
Local translation in neurons: visualization and function
Nature Structural & Molecular Biology (2019)
-
Intron retention and nuclear loss of SFPQ are molecular hallmarks of ALS
Nature Communications (2018)