Abstract
Precise regulation of transcription is crucial for the cellular mechanisms underlying memory formation. However, the link between neuronal stimulation and the proteins that directly interact with histone modifications to activate transcription in neurons remains unclear. Brd4 is a member of the bromodomain and extra-terminal domain (BET) protein family, which binds acetylated histones and is a critical regulator of transcription in many cell types, including transcription in response to external cues. Small molecule BET inhibitors are in clinical trials, yet almost nothing is known about Brd4 function in the brain. Here we show that Brd4 mediates the transcriptional regulation underlying learning and memory. The loss of Brd4 function affects critical synaptic proteins, which results in memory deficits in mice but also decreases seizure susceptibility. Thus Brd4 provides a critical link between neuronal activation and the transcriptional responses that occur during memory formation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Hsieh, J. & Gage, F.H. Chromatin remodeling in neural development and plasticity. Curr. Opin. Cell Biol. 17, 664–671 (2005).
Gapp, K., Woldemichael, B.T., Bohacek, J. & Mansuy, I.M. Epigenetic regulation in neurodevelopment and neurodegenerative diseases. Neuroscience 264, 99–111 (2014).
Frey, U., Frey, S., Schollmeier, F. & Krug, M. Influence of actinomycin D, a RNA synthesis inhibitor, on long-term potentiation in rat hippocampal neurons in vivo and in vitro. J. Physiol. (Lond.) 490, 703–711 (1996).
Frey, U., Krug, M., Brödemann, R., Reymann, K. & Matthies, H. Long-term potentiation induced in dendrites separated from rat's CA1 pyramidal somata does not establish a late phase. Neurosci. Lett. 97, 135–139 (1989).
Nguyen, P.V., Abel, T. & Kandel, E.R. Requirement of a critical period of transcription for induction of a late phase of LTP. Science 265, 1104–1107 (1994).
Messaoudi, E., Ying, S.-W., Kanhema, T., Croll, S.D. & Bramham, C.R. Brain-derived neurotrophic factor triggers transcription-dependent, late phase long-term potentiation in vivo. J. Neurosci. 22, 7453–7461 (2002).
Sims, R.J., Belotserkovskaya, R. & Reinberg, D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18, 2437–2468 (2004).
Kadonaga, J.T. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116, 247–257 (2004).
Jang, M.K. et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19, 523–534 (2005).
Liu, W. et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155, 1581–1595 (2013).
Zippo, A. et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138, 1122–1136 (2009).
Nagarajan, S. et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 8, 460–469 (2014).
Belkina, A.C. & Denis, G.V. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer 12, 465–477 (2012).
Wu, S.-Y., Lee, A.-Y., Lai, H.-T., Zhang, H. & Chiang, C.-M. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol. Cell 49, 843–857 (2013).
Houzelstein, D. et al. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22, 3794–3802 (2002).
Descalzi, G., Fukushima, H., Suzuki, A., Kida, S. & Zhuo, M. Genetic enhancement of neuropathic and inflammatory pain by forebrain upregulation of CREB-mediated transcription. Mol. Pain 8, 90 (2012).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Hargreaves, D.C., Horng, T. & Medzhitov, R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138, 129–145 (2009).
Saha, R.N. et al. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat. Neurosci. 14, 848–856 (2011).
Fischer, A., Sananbenesi, F., Mungenast, A. & Tsai, L.-H. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol. Sci. 31, 605–617 (2010).
Schael, S. et al. Casein kinase 2 phosphorylation of protein kinase C and casein kinase 2 substrate in neurons (PACSIN) 1 protein regulates neuronal spine formation. J. Biol. Chem. 288, 9303–9312 (2013).
Delmore, J.E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Mirguet, O. et al. Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J. Med. Chem. 56, 7501–7515 (2013).
Asangani, I.A. et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510, 278–282 (2014).
Kanno, T. et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat. Struct. Mol. Biol. 21, 1047–1057 (2014).
Lussier, M.P., Gu, X., Lu, W. & Roche, K.W. Casein kinase 2 phosphorylates GluA1 and regulates its surface expression. Eur. J. Neurosci. 39, 1148–1158 (2014).
Sanz-Clemente, A., Matta, J.A., Isaac, J.T.R. & Roche, K.W. Casein kinase 2 regulates the NR2 subunit composition of synaptic NMDA receptors. Neuron 67, 984–996 (2010).
Dietrich, J.-B. The MEF2 family and the brain: from molecules to memory. Cell Tissue Res. 352, 179–190 (2013).
Korb, E. & Finkbeiner, S. Arc in synaptic plasticity: from gene to behavior. Trends Neurosci. 34, 591–598 (2011).
Borges, K. & Dingledine, R. Functional organization of the GluR1 glutamate receptor promoter. J. Biol. Chem. 276, 25929–25938 (2001).
Myers, S.J. et al. Transcriptional regulation of the GluR2 gene: neural-specific expression, multiple promoters, and regulatory elements. J. Neurosci. 18, 6723–6739 (1998).
Matzuk, M.M. et al. Small-molecule inhibition of BRDT for male contraception. Cell 150, 673–684 (2012).
Antunes, M. & Biala, G. The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn. Process. 13, 93–110 (2012).
Naydenov, A.V. et al. ABHD6 blockade exerts antiepileptic activity in PTZ-induced seizures and in spontaneous seizures in R6/2 mice. Neuron 83, 361–371 (2014).
Ferraro, T.N. et al. Mapping loci for pentylenetetrazol-induced seizure susceptibility in mice. J. Neurosci. 19, 6733–6739 (1999).
Dhir, A. Pentylenetetrazol (PTZ) kindling model of epilepsy. Curr. Protoc. Neurosci. Ch. 9, unit 9.37 (2012).
Bialer, M. & White, H.S. Key factors in the discovery and development of new antiepileptic drugs. Nat. Rev. Drug Discov. 9, 68–82 (2010).
Brehme, H., Kirschstein, T., Schulz, R. & Köhling, R. In vivo treatment with the casein kinase 2 inhibitor 4,5,6,7- tetrabromotriazole augments the slow afterhyperpolarizing potential and prevents acute epileptiform activity. Epilepsia 55, 175–183 (2014).
Rogawski, M.A. AMPA receptors as a molecular target in epilepsy therapy. Acta Neurol. Scand. Suppl. 2013, 9–18, (2013).
Zhang, J. & Abdullah, J.M. The role of GluA1 in central nervous system disorders. Rev. Neurosci. 24, 499–505 (2013).
Kato, A.S., Gill, M.B., Yu, H., Nisenbaum, E.S. & Bredt, D.S. TARPs differentially decorate AMPA receptors to specify neuropharmacology. Trends Neurosci. 33, 241–248 (2010).
Yamaguchi, S., Donevan, S.D. & Rogawski, M.A. Anticonvulsant activity of AMPA/kainate antagonists: comparison of GYKI 52466 and NBOX in maximal electroshock and chemoconvulsant seizure models. Epilepsy Res. 15, 179–184 (1993).
Namba, T., Morimoto, K., Sato, K., Yamada, N. & Kuroda, S. Antiepileptogenic and anticonvulsant effects of NBQX, a selective AMPA receptor antagonist, in the rat kindling model of epilepsy. Brain Res. 638, 36–44 (1994).
Hara, H. et al. Effect of YM872, a selective and highly water-soluble AMPA receptor antagonist, in the rat kindling and rekindling model of epilepsy. Eur. J. Pharmacol. 531, 59–65 (2006).
Kodama, M. et al. Effects of YM90K, a selective AMPA receptor antagonist, on amygdala-kindling and long-term hippocampal potentiation in the rat. Eur. J. Pharmacol. 374, 11–19 (1999).
Mattes, H., Carcache, D., Kalkman, H.O. & Koller, M. α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) antagonists: from bench to bedside. J. Med. Chem. 53, 5367–5382 (2010).
Rakhade, S.N. et al. Glutamate receptor 1 phosphorylation at serine 831 and 845 modulates seizure susceptibility and hippocampal hyperexcitability after early life seizures. J. Neurosci. 32, 17800–17812 (2012).
Banaszynski, L.A. et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013).
Acknowledgements
We thank K. Ozato for the Brd4 construct, J. Gresack and the Rockefeller behavioral core for guidance and suggestions, K. Thomas for microscopy expertise, the Memorial Sloan-Kettering Cancer Center histology core for immunohistochemistry support, A. Soshnev for the schematic model graphic, J. Gerace for editing and members of the Allis laboratory for feedback. Support for this work was provided by a grant from the Robertson Foundation, a Ruth Kirschstein US National Research Service Award fellowship (F32MH103921), the US National Institutes of Health (R01 NS34389, NS081706) and a Simons Foundation Research Award. R.B.D. is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
E.K. wrote the manuscript and designed and carried out experiments. M.H. helped carry out behavioral testing and molecular studies. I.Z.-S. helped with seizure testing. R.B.D. helped initiate and design the project and provided feedback, and C.D.A. provided support, feedback and guidance.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
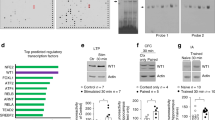
Supplementary Figure 1 Brd4 is expressed in the brain.
(a) Whole cell lysate of regions of adult mouse brain. Representative blot of 3 biological replicates. (b, c) Cultured cortical neurons stained with Brd4 and CamKII (b) or Gaba (c). (d) Brd4 mRNA from cultured neurons after BDNF treatment (one sample t-test for 0.5 hours n = 14, for 2 hours n = 13, for 4 hours n = 14, for 8 hours n = 12, for 24 hours n = 13, P = 0.0339, t = 2.01). (e, f) Brd4 protein after short (e) or long (f) BDNF treatment. Representative blot of 3 biological replicates. *, p<0.05. Full-length blots are presented in Supplementary Figure 10. min, minutes. h, hours. Error bars represent standard error. Scale bar is 10 μm.
Supplementary Figure 2 Brd4 regulates IEG transcription in response neurons.
(a, b, c) Time course of the BDNF-induced increase in Arc (a), Fos (b) and Nr4a1 (c) after (-) or (+)Jq1 treatment. (d) Brd4 mRNA 3 days after lentiviral infection of an siRNA targeted against Brd4 compared to a scrambled siRNA (one-sample t test, n = 7 biological replicates P = 6.18E-6, t = 13.07). (e) BDNF-induced MAPK phosphorylation after Brd4 knockdown with lentiviral infection. Representative of 2 biological replicates. (f, g) Staining (f) and quantification (g) of Brd4 in neurons 5 days after transfection with GFP and Brd4 siRNA (unpaired two-sided t test n = 45 neurons for control siRNA and 37 neurons for Brd4 siRNA from 5 biological replicates, P = 6.38E-16, t = 10.093). (h, i) Brd2 (h) and Brd3 (i) mRNA fold decrease 5 days after transfection in N2A cells (one sample t-test n = 3 biological replicates, for Brd2 P = 0.0157, t = 5.51, for Brd3 P = 0.019, t = 4.92). (i) Quantification of Brd4 staining 5 days after transfection with different siRNAs targeted against Brd4. N = 47 neurons for control siRNA (upaired two-tailed t test for control n = 47 for Brd4 siRNA1 n = 43 and for Brd4 siRNA2 n = 37 neurons from 2 biological replicates, for siRNA1 P = 2.24E-38, t = 22.91, for siRNA2 P = 2.82E-25, t = 15.18). (k, l) Staining (k) and quantification (l) of Arc with or without 30 minutes BDNF stimulation after transfection with GFP and either a nontargeting siRNA or different Brd4 siRNA constructs (upaired two-tailed t test for control n = 25, for control + BDNF n = 23, for Brd4 siRNA1 n = 22, for Brd4 siRNA1 + BDNF n = 22, for Brd4 siRNA2 n = 14, and for Brd4 siRNA2 + BDNF n = 26 from 2 biological replicates, for control vs BDNF P = 9.53E-4, t = 3.432). (m) Quantification of Arc staining in neurons transfected with a long or short form of Brd4 (upaired two-tailed t test for control n = 42 neurons, for long Brd4 n = 26 neurons and for short Brd4 n = 30 neurons, for control vs long Brd4 P = 0.015, t = 2.48, for long vs short Brd4 P = 0.0016, t = 3.29). (n) Decrease in Arc, Fos and Nr4a1 RNA after 24 hours of Jq1 treatment in the absence of exogenous stimulation in cultured cortical neurons (one sample t test, for Arc n = 13 biological replicates P = 3.098E-5, t = 6.58, for Fos n = 13 biological replicates P = 0.013, t = 2.65, and for Nr4a1 n = 11, P = 0.00104, t = 4.75). (o) Jq1 does not affect global H3 or H4 acetyl levels. (p) MAPK phosphorylation in response to BDNF with or without Jq1 pretreatment. Representative blot of 2 biological replicates. a.u., arbitrary units. #, p< 0.05 for univariate analysis. ###, p< 0.001 for univariate analysis. *, p< 0.05. ***, p<0.001. Full-length blots are presented in Supplementary Figure 10. Error bars represent standard error. Scale bar is 10 μM.
Supplementary Figure 3 RNA-sequencing analysis of effects of Jq1 in neurons.
(a) Experimental paradigm for genome-wide analysis of effects of Jq1. (b) Heat map of fold induction of all genes significantly induced by BDNF after treatment with (-) or (+) Jq1. (c) Top GO terms of genes clusters enriched in Jq1 up-regulated genes. Data represents averages of 3 biological replicates. (d) Histone acetylation at promoter regions of IEGs in response to BDNF for acetyl marks that recruit Brd4. N = 3 biological replicates. (e) Western blot analysis of acetylated histone H4K16 and H3K14 with (-) or (+) Jq1. Representative blot of 3 biological replicates. Full-length blots are presented in Supplementary Figure 11. min, minutes.
Supplementary Figure 4 CK2 regulates Brd4 in neurons.
(a, b, c) Brd4 ChIP-qPCR analysis in neurons stimulated with BDNF to measure Brd4 at promoters of Arc (a), Fos (b), and Nr4a1 (c). (d, e, f) Brd2 ChIP in neurons stimulated with BDNF at promoters of Arc (d), Fos (e), and Nr4a1 (f). N = 3 biological replicates. (g, h, i) Brd3 ChIP in neurons stimulated with BDNF at promoters of Arc (g), Fos (h), and Nr4a1 (i). N = 3 biological replicates for a-i. (j) MAPK phosphorylation in response to BDNF in the presence of TBB. Representative of 3 biological replicates. (k, l, m) CBP ChIP-qPCR analysis in neurons stimulated with BDNF with Jq1 or TBB pretreatment to measure Brd4 at promoters of Arc (k), Fos (l), and Nr4a1 (m). N = 2 biological replicates. (n) Quantification of Ck2 expression in neurons transfected with GFP and either nontargeting siRNA, or siRNA targeting CK2 (unpaired two-tailed t test, for control siRNA n = 24 neurons and for CK2 siRNA n = 26 neurons from 3 biological replicates, P = 8.05E-5, t = 3.29). (o, p) Arc and CK2 staining (o) and Arc quantification (p) after a 30 minute BDNF stimulation of neurons transfected with GFP and either a nontargeting siRNA pool or a siRNA pool targeting CK2 (unpaired two-tailed t test, for control siRNA n = 50 neurons, n = 39 for BDNF, for CK2 siRNA n = 39, and for CK2 siRNA + BDNF n = 37 from 5 biological replicates, control vs BDNF, P = 1.21E-5, t = 5.244, for BDNF vs CK2 siRNA + BDNF P = 7.09E-7, t = 5.42). (q) Mobile fraction quantification of EGFP-Brd4 (unpaired two-tailed t test for control n = 15 for BDNF 18 neurons. P = 5.389E-5, t = 4.73). ***, p<0.001. a.u. arbitrary units. a.u. arbitrary units. min, minutes. Error bars represent standard error. Full-length blots are presented in Supplementary Figure 11. Scale bar is 10 μm.
Supplementary Figure 5 Phosphorylation of the CK2 site in Brd4.
(a) Dot blot for phopho-Brd4 antisera using target and control peptides shows specific binding to peptides containing phosphorylated S492. (b) Western blot with phopho-Brd4 antisera in lyates treated with phosphatase or control lysates. Blots are representative of 3 replicates. (c) Quantification of Brd4 expression shows elevated and equivalent Brd4 levels in neurons transfected with GFP and Brd4 with deletions or mutations in the CK2 site (unpaired two-sided t test, for GFP n = 68, for Brd4 n = 61, P = 4.596E-17, t = 9.73, for CK2 deletion n = 46, P = 1.26E-27, t = 14.57 for deletion 492-494 n = 44, P = 1.48E-22, t = 12.37, for S492A n = 54, P = 4.14E-34, t = 17.17, for SSS492ESE n = 51 from 5 biological replicates, P = 7.82E-24, t = 12.73, no significant differences between Brd4 constructs). (d) Example images of EGFP-Brd4 with CK2 site mutations costained with H4K16acetyl. (e) Example graphs showing corresponding changes in fluorescent signal of Brd4 and H4K16acetyl in sections of neuronal nuclei. **, p<0.01. ***, p<0.001. Full-length blots are presented in Supplementary Figure 11. Error bars represent standard error. Scale bar is 5 μm.
Supplementary Figure 6 Brd4 inhibition decreases surface GluA1.
(a) Quantification of mRNA after different times of Jq1 treatment. N = 3 biological replicates for Arc and Fos, N = 8 for Nr4a1, N = 9 for Gria1, N = 4 for Bdnf and N = 5 for Brd4. (b) Quantification of hippocampal neurons treated with Jq1 for 24 hours and stained for surface GluA1 (unpaired two-sided t test, for control n = 13 neurons and for Jq1 n = 12 neurons from 2 biological replicates, P = 0.0015, t = 3.56). (c, d) Images (c) and quantification (d) of surface GluA1 staining 5 days after transfection with different siRNAs targeted against Brd4 (unpaired two-sided t test, for control siRNA n = 14 neurons, for Brd4 siRNA1 n = 13 neurons, and for Brd4 siRNA2 n = 14 neurons from 2 biological replicates, for control vs siRNA1 P = 0.0114, t = 2.61, for control vs siRNA2 P = 0.0078, t = 2.76). (e) Quantification of spine number in neurons treated with Jq1 for 24 hours (unpaired two-sided t test, n = 68 neurons for control for Jq1 and n = 66 from 2 biological replicates). (f) Quantification of staining of surface GluA1 in neurons with long or short forms of Brd4 (unpaired two-sided t test, for control n = 15 neurons, for long Brd4 n = 17, and for short Brd4 n = 18 from 2 biological replicates, for control vs long Brd4 P = 0.0161, t = 2.55, for long vs short Brd4 P = 0.0308, t = 2.26). (g) ChIP for Brd4 at the gria1 promoter region. N = 7 biological replicates for control conditions, 3 biological replicates for Jq1 conditions. (h) ChIPs for histone acetylation at the gria1 promoter region. N = 2 biological replicates. *, p<0.05. ***, p<0.001. a.u. arbitrary units. n.s., non-significant. Scale bar is 10 μm.
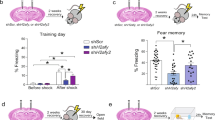
Supplementary Figure 7 Jq1 affects mouse behavior.
(a) Weight change in mice treated daily with Jq1 or vehicle. (b) Zone preference ratio in an open field in mice treated with vehicle or with Jq1 for 1 or 3 weeks. (c) Total distance traveled during 1 hour of an open field in mice treated with vehicle or with Jq1 for 1 or 3 weeks. (d) Time spent moving during 1 hour of an open field in mice treated with vehicle or with Jq1 for 1 or 3 weeks. (e) Distance traveled in the novel object box during habituation. (f-i) Time spent exploring objects for memory tests during initial exposure (f) or during testing (g) for memory tests or for learning tests during initial exposure (h) or during testing (i). (j) Discrimination index of time spent with a novel vs familiar object when mice are tested immediately after initial exposure to objects (for learning tests, n = 10 mice for DMSO and 9 mice for 1 week Jq1. No significant difference between DMSO and Jq1, for one sample t test for DMSO P = 0.00325, t = 4.14, for Jq1 P = 0.00103, t = 5.017). (k) Percent of time spent freezing in response to a tone before training, during training, or 1 day after training in a new context (2-way ANOVA with posthoc t test, P = 2E-16, F = 165.7, df = 130) (l) Percent of time spent freezing before, immediately after, or 1 day after fear conditioning training in the training context or in a novel context. (2-way ANOVA with posthoc t test, P = 2E-16, F = 80.72, df = 110). N = 10 mice for control and 3 week Jq1 and 9 mice for 1 week Jq1 for testing groups. *, p<0.05. ***, p<0.001. n.s., nonsignificant. Error bars represent standard error.
Supplementary Figure 8 Jq1 affects seizure susceptibility.
(a) Quantification of mRNA from cortical tissue from mice after behavioral testing following treatment with DMSO or 1 week of Jq1 (unpaired two-sided t test for dmso n = 9 brains and for Jq1 n = 8, for gria1 P = 0.00517, t value = 3.27, for Nr4a1 P = 0.018, t value = 2.64). (b) Western of GluA1 levels after Jq1 treatment. (c) Data for each level of Racine scoring of seizure induction for mice treated for one week with either DMSO or Jq1 and then given pentylenetetrazol (PTZ) to induce seizures. (d) Percent mortality of male mice after seizure injection by PTZ. (e) Seizure susceptibility score of female mice treated for one week with either DMSO or Jq1 and given pentylenetetrazol (PTZ) to induce seizures. (f) Percent mortality of female mice after seizure injection by PTZ. (g, h) Seizure susceptibility scores during kindling for individual mice treated with DMSO (f) or Jq1 (g). N = 9 mice per group. *, p<0.05. ***, p<0.001. Error bars represent standard error. Full-length blots are presented in Supplementary Figure 11.
Supplementary Figure 9 Model of Brd4 function in neurons.
Neuronal activation by signaling molecules such as BDNF trigger activation of CK2 which in turn phosphorylates Brd4. Brd4 binds to acetylated lysines on histone proteins and activates expression of neuronal genes including IEGs which then affect synaptic proteins. Inhibition of Brd4 by Jq1 affects aspects synaptic function and memory formation.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 (PDF 2860 kb)
Rights and permissions
About this article
Cite this article
Korb, E., Herre, M., Zucker-Scharff, I. et al. BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat Neurosci 18, 1464–1473 (2015). https://doi.org/10.1038/nn.4095
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4095
This article is cited by
-
Remodeling of the postsynaptic proteome in male mice and marmosets during synapse development
Nature Communications (2024)
-
Inhibition of CK2 Diminishes Fibrotic Scar Formation and Improves Outcomes After Ischemic Stroke via Reducing BRD4 Phosphorylation
Neurochemical Research (2024)
-
The 100 most-cited manuscripts in epilepsy epigenetics: a bibliometric analysis
Child's Nervous System (2023)
-
Effect of BRD4 Inhibitor on Cognitive Deficit and c-Fos /BDNF level in rats with Alzheimer's disease
Neuroscience and Behavioral Physiology (2023)
-
Scrutinizing the Therapeutic Potential of PROTACs in the Management of Alzheimer’s Disease
Neurochemical Research (2023)