Abstract
Lineage progression and diversification is regulated by the coordinated action of unique sets of transcription factors. Oligodendrocytes (OL) and astrocytes (AS) comprise the glial sub-lineages in the CNS, and the manner in which their associated regulatory factors orchestrate lineage diversification during development and disease remains an open question. Sox10 and NFIA are key transcriptional regulators of gliogenesis associated with OL and AS. We found that NFIA inhibited Sox10 induction of OL differentiation through direct association and antagonism of its function. Conversely, we found that Sox10 antagonized NFIA function and suppressed AS differentiation in mouse and chick systems. Using this developmental paradigm as a model for glioma, we found that this relationship similarly regulated the generation of glioma subtypes. Our results describe the antagonistic relationship between Sox10 and NFIA that regulates the balance of OL and AS fate during development and demonstrate for the first time, to the best of our knowledge, that the transcriptional processes governing glial sub-lineage diversification oversee the generation of glioma subtypes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kang, P. et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the onset of gliogenesis. Neuron 74, 79–94 (2012).
Deneen, B. et al. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52, 953–968 (2006).
Namihira, M. et al. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev. Cell 16, 245–255 (2009).
Fancy, S.P. et al. Evidence that nuclear factor IA inhibits repair after white matter injury. Ann. Neurol. 72, 224–233 (2012).
Stiles, C.D. & Rowitch, D.H. Glioma stem cells: a midterm exam. Neuron 58, 832–846 (2008).
de la Iglesia, N. et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 22, 449–462 (2008).
Ligon, K.L. et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron 53, 503–517 (2007).
Glasgow, S.M. et al. The miR-223/nuclear factor I-A axis regulates glial precursor proliferation and tumorigenesis in the CNS. J. Neurosci. 33, 13560–13568 (2013).
Maher, E.A. et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 15, 1311–1333 (2001).
Stolt, C.C. et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 16, 165–170 (2002).
Kuhlbrodt, K. et al. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 18, 237–250 (1998).
Zhou, Q., Wang, S. & Anderson, D.J. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron 25, 331–343 (2000).
Li, H., Lu, Y., Smith, H.K. & Richardson, W.D. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J. Neurosci. 27, 14375–14382 (2007).
Liu, Z. et al. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev. Biol. 302, 683–693 (2007).
Pedraza, C.E. et al. Production, characterization and efficient transfection of highly pure oligodendrocyte precursor cultures from mouse embryonic neural progenitors. Glia 56, 1339–1352 (2008).
Gabay, L., Lowell, S., Rubin, L. & Anderson, D.J. Deregulation of dorsoventral pattering by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron 40, 485–499 (2003).
Tsai, H.H. et al. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science 337, 358–362 (2012).
Masahira, N. et al. Olig2-positive progenitors in the embryonic spinal cord give rise not only to motoneurons and oligodendrocytes, but also to a subset of astrocytes and ependymal cells. Dev. Biol. 293, 358–369 (2006).
Matsuoka, T. et al. Neural crest origins of the neck and shoulder. Nature 436, 347–355 (2005).
Britsch, S. et al. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 15, 66–78 (2001).
Hochstim, C., Deneen, B., Lukaszewicz, A., Zhou, Q. & Anderson, D.J. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell 133, 510–522 (2008).
Bannykh, S.I., Stolt, C.C., Kim, J., Perry, A. & Wegner, M. Oligodendroglial-specific transcriptional factor SOX10 is ubiquitously expressed in human gliomas. J. Neurooncol. 76, 115–127 (2006).
Rousseau, A. et al. Expression of oligodendroglial and astrocytic lineage markers in diffuse gliomas: use of YKL-40, ApoE, ASCL1, and NKX2–2. J. Neuropathol. Exp. Neurol. 65, 1149–1156 (2006).
Ferletta, M., Uhrbom, L., Olofsson, T., Ponten, F. & Westermark, B. Sox10 has a broad expression pattern in gliomas and enhances platelet-derived growth factor-B–induced gliomagenesis. Mol. Cancer Res. 5, 891–897 (2007).
Song, H.R. et al. Nuclear factor IA is expressed in astrocytomas and is associated with improved survival. Neuro-oncol. 12, 122–132 (2010).
Liu, A. et al. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 25, 4833–4842 (2006).
Heng, Y.H. et al. NFIX regulates neural progenitor cell differentiation during hippocampal morphogenesis. Cereb. Cortex 24, 261–279 (2012).
Deneen, B. et al. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52, 953–968 (2006).
Cai, J. et al. A crucial role for Olig2 in white matter astrocyte development. Development 134, 1887–1899 (2007).
Ono, K. et al. Regional- and temporal-dependent changes in the differentiation of Olig2 progenitors in the forebrain, and the impact on astrocyte development in the dorsal pallium. Dev. Biol. 320, 456–480 (2008).
Zhu, X., Bergles, D. & Nishiyama, A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development 135, 145–157 (2008).
Zhu, X., Hill, R.A. & Nishiyama, A. NG2 cells generate oligodendrocytes and gray matter astrocytes in the spinal cord. Neuron Glia Biol. 4, 19–26 (2008).
Holland, E.C. et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat. Genet. 25, 55–57 (2000).
Dai, C. et al. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 15, 1913–1925 (2001).
Morgan, B.A. & Fekete, D. Manipulating gene expression with replication-competent retroviruses. Methods Cell Biol. 51, 185–218 (1996).
Momose, T. et al. Efficient targeting of gene expression in chick embryos by microelectroporation. Dev. Growth Differ. 41, 335–344 (1999).
Wissmüller, S., Kosian, T., Wolf, M., Finzsch, M. & Wegner, M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 34, 1735–1744 (2006).
Pedraza, C.E., Monk, R., Lei, J., Hao, Q. & Macklin, W.B. Production, characterization, and efficient transfection of highly pure oligodendrocyte precursor cultures from mouse embryonic neural progenitors. Glia 56, 1339–1352 (2008).
Chen, F. & LoTurco, J. A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J. Neurosci. Methods 207, 172–180 (2012).
Acknowledgements
We thank A. Groves for assistance with the GST pull-down studies. This work was supported by grants from the National Multiple Sclerosis Society (RG 4623A1/2 to B.D.), the Sontag Foundation (B.D.), and the US National Institutes of Health (R01 NS071153 to B.D., R01 HD062553 to J.L.N., R01HD055655 and R01MH056524 to J.J.L., 5-T32HL092332-08 to S.G.). The GFAP-Luc reporter was a kind gift from M. Brenner (University of Alabama-Birmingham).
Author information
Authors and Affiliations
Contributions
B.D. and S.M.G. conceived the project, designed the experiments and wrote the manuscript. S.M.G. and W.Z. performed all of the experiments. B.D., S.M.G., W.Z. and C.M. analyzed the data. C.C.S., M.W., T.-W.H., J.L.N., J.J.L. and F.C. provided essential reagents.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 NFIA and Sox10 expression during OLP development
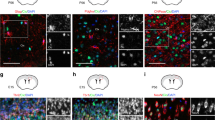
NFIA is co-expressed with Sox10 in the spinal cord at E12.5 (B). NFIA is expressed in OLPs as demonstrated by co-expression with Sox10 (C, filled arrows) at E18.5, but not markers of mature OLs MBP (D, E) or PLP (F, G), denoted by unfilled arrows. These expression dynamics are summarized schematically in A. Sox10 is co-expressed with Glast::dsRed (H) and Olig2 (I) in the VZ at E12.5.
Supplementary Figure 2 NFIA antagonizes ectopic induction of OLP markers by Sox10.
Overexpression of NFIA (A), Sox10 (B), or Sox10+NFIA (C) in the chick spinal cord. Sections are adjacent or near adjacent to the ones presented in Figure 1A-H. Arrow in B denotes ectopic Olig2 expression, while arrow in C denotes absence of Olig2. Graph in E represents analysis of Caspase 3 staining in adjacent sections. Image in F is from Figure 1F, boxed region is shown at higher magnification in G to demonstrate overlapping expression of Sox10 and NFIA. (H) Quantification of Sox10 expression using fluorescence intensity on chick spinal cord sections stained with α-HA to detect ectopic Sox10 expression in these studies. (I-J) Cultured OLPs infected with NFIA or GFP control viruses prior to mitogen removal and differentiation, stained with the OLP marker, Nkx2.2 Arrows denote viral infected, Nkx2.2-expressing OLP populations. Quantification in D-E and H is from 8 independent spinal cords, 8 sections/spinal cord.
Supplementary Figure 3 NFI and Sox binding sites are present in myelin and astrocyte gene promoters
Schematic representation of myelin gene promoters (MBP, PLP, MAG) and astrocyte gene promoters (GFAP, APCDD1) depicting the location of Sox and NFI sites. Also shown is the conservation of these sites between human and mouse promoters.
Supplementary Figure 4 Single-channel images from Figure 3s–u.
A, GFP control, B, NFIA transfection, Sox10 transfection. Filled arrows denote overlap between GFP and GFAP, Open arrow denote non-overlapping GFP and GFAP expression. The extent of GFAP/GFP overlap is increased in the NFIA transfections (B) and decreased in the Sox10 transfections (C), compared to the control sample (A). See Figure 3W for quantification.
Supplementary Figure 5 Sox10-positive cells give rise to gray- and white-matter astrocytes
The Sox10-Cre;Rosa26-LacZ mouse line was used to trace the fate of Sox10-expressing cells in the spinal cord from P0-P1 neonatal mice. Sox10 expressing cells give rise to AldoC- (A), GFAP-(B), S100β-(C), and GS-(D) expressing white matter astrocytes (A-C) and grey-matter astrocytes (A, C-D).
Supplementary Figure 6 Sox10LacZ/LacZ neural stem cells demonstrate increased astrocyte production.
Neural stem cells from Sox10LacZ/+ and Sox10LacZ/LacZ E13.5 spinal cord were infected with dsRed control (A-B) or Sox10-dsRed (C) retrovirus and allowed to differentiation. Five days after plating on adherent substrate, cultures were fixed and stained with GFAP. Quantification in D is derived from four independent experiments, on two independently derived sets of neural stem cells from Sox10LacZ/+ and Sox10LacZ/LacZ litters. In D, Het is Sox10LacZ/+ and KO is Sox10LacZ/LacZ. *p<0.001. All error bars represent SD.
Supplementary Figure 7 Loss of Sox10 does not result in an increase in white matter astrocytes
In the absence of Sox10 no change in the number of NKX6.1+/NFIA-expressing cells is observed (A-C, filled arrows). In addition, no difference was observed in the number of β-gal/NKX6.1 or β-gal/Pax6 WMAs in the absence of Sox10 (D-I, unfilled arrows). In C, F, I, “Het” corresponds to Sox10LacZ/+ and “KO” corresponds to Sox10LacZ/LacZ. Analysis of Ki-67 in Sox10LacZ/+ and Sox10LacZ/LacZ embryos did not reveal any changes in proliferation (J-L). All quantifications are derived from three embryos of each genotype from two independent litters.
Supplementary Figure 8 Generation of oligodendroglioma using IUE and PiggyBacs to target OL lineages with oncogenes
Brain from P14 mouse IUE at E16.5 with GLAST-PBase and MBP-GFPt2aRas (A). Arrow denotes region electroporated and dotted line denotes cross section analyzed in B; dotted line in B is midlne. H&E staining of tumors (C-D). Tumors demonstrate pathological hallmarks of oligodendrolglioma, including small monomorphic nuclei lacking processes, perineuronal satellitosis, and invasion into perivascular and subpial spaces. Dashed box in C is magnified area in D. Circle in D demonstrates typical perineuronal satellitosis. Olig2 staining in mouse oligodendroglioma (E-H). Co-expression of ectopic HA-NFIA with Ras-GFP expressing cells in astrocytoma-like tumors (H-J).
Supplementary Figure 10 Uncropped gel and blot images.
(A) Reprint of Figure 5E with individual blots labeled 1-7 (ie. αFlag-1, etc.). (B) Corresponding uncropped blots of those shown in “A” and Figure 5E. Uncropped blots are the size of the membrane which was blotted. Membranes were often cut at or below 50kDa to enable blotting for multiple antibodies. Representative blots WB α-Flag-1 and WB α-Olig2-2 were taken from the same membrane, where the dashed line represents where the membrane was cut. Lanes are numbered under the blot image and L indicates the ladder lane. The protein standards are depicted to the right of the blots. (C) Uncropped image of exposed GST pulldown assay and western blot. (D) Uncropped blots of Co-IP shown in Figure 2A. The protein standards are depicted to the right of the blots. (E) Uncropped images of DNA gels for ChIP analysis for experiments performed in Figures 2B,C,D and 3L,M.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 (PDF 2700 kb)
Supplementary Methods Checklist
(PDF 383 kb)
Rights and permissions
About this article
Cite this article
Glasgow, S., Zhu, W., Stolt, C. et al. Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci 17, 1322–1329 (2014). https://doi.org/10.1038/nn.3790
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3790
This article is cited by
-
Understanding current experimental models of glioblastoma-brain microenvironment interactions
Journal of Neuro-Oncology (2024)
-
IFN-γ-STAT1-mediated CD8+ T-cell-neural stem cell cross talk controls astrogliogenesis after spinal cord injury
Inflammation and Regeneration (2023)
-
Analysis of cnidarian Gcm suggests a neuronal origin of glial EAAT1 function
Scientific Reports (2023)
-
Comprehensive analysis of microglia gene and subpathway signatures for glioma prognosis and drug screening: linking microglia to glioma
Journal of Translational Medicine (2022)
-
Human transcription factor protein interaction networks
Nature Communications (2022)