Abstract
MATR3 is an RNA- and DNA-binding protein that interacts with TDP-43, a disease protein linked to amyotrophic lateral sclerosis (ALS) and frontotemporal dementia. Using exome sequencing, we identified mutations in MATR3 in ALS kindreds. We also observed MATR3 pathology in ALS-affected spinal cords with and without MATR3 mutations. Our data provide more evidence supporting the role of aberrant RNA processing in motor neuron degeneration.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Renton, A.E., Chiò, A. & Traynor, B.J. Nat. Neurosci. 17, 17–23 (2014).
Feit, H. et al. Am. J. Hum. Genet. 63, 1732–1742 (1998).
Senderek, J. et al. Am. J. Hum. Genet. 84, 511–518 (2009).
Eisen, A. & Kuwabara, S. J. Neurol. Neurosurg. Psychiatry 83, 399–403 (2012).
Ling, S.C. et al. Proc. Natl. Acad. Sci. USA 107, 13318–13323 (2010).
Salton, M. et al. PLoS ONE 6, e23882 (2011).
Cookson, M.R. Biochem. Soc. Trans. 40, 1070–1073 (2012).
van Blitterswijk, M. et al. PLoS ONE 8, e60788 (2013).
Zhang, Z. & Carmichael, G.G. Cell 106, 465–475 (2001).
Höck, J. et al. EMBO Rep. 8, 1052–1060 (2007).
Ma, H., Siegel, A.J. & Berezney, R. J. Cell Biol. 146, 531–542 (1999).
Giordano, G. et al. J. Neurochem. 94, 808–818 (2005).
Aliaga, L. et al. Hum. Mol. Genet. 22, 4293–4305 (2013).
Chiò, A. et al. Amyotroph. Lateral Scler. 10, 310–323 (2009).
Johnson, J.O. et al. Neuron 68, 857–864 (2010).
Kim, H.J. et al. Nature 495, 467–473 (2013).
Weihl, C.C. et al. J. Neurol. Neurosurg. Psychiatry 79, 1186–1189 (2008).
Acknowledgements
DNA samples for this study were obtained in part from the National Institute of Neurological Disorders and Stroke (NINDS) repository at the Coriell Cell Repositories (http://www.coriell.org/). We thank the patients and research subjects who contributed samples for this study. This work was supported in part by the Intramural Research Programs of the US National Institutes of Health (NIH), National Institute on Aging (Z01-AG000949-02) and NINDS. The work was also supported by the Packard Center for ALS Research at Johns Hopkins (B.J.T.), ALS Association (B.J.T., A. Chiò), Ontario Research Fund (E.R.), UK MND Association (J.H., R.W.O. grant 11/6075), Medical Research Council (MRC) UK (J.H.), Wellcome Trust/MRC Joint Call in Neurodegeneration Award (J.H., grant WT089698), MRC Neuromuscular Centre (J.H.), UK National Institute for Health Research Biomedical Research Unit (J.H.), Biomedical Research Centre (A. Pittman), MRC/Motor Neuron Disease Association Lady Edith Wolfson fellowship (P.F.), AriSLA — Italian Research Foundation for Amyotrophic Lateral Sclerosis (A. Chiò, B.J.T.), Italian Health Ministry (Ricerca Sanitaria Finalizzata 2007, A. Chiò), Fondazione Vialli e Mauro Onlus (A. Chiò), Federazione Italiana Giuoco Calcio (A. Chiò, M. Sabatelli, B.J.T.), Compagnia di San Paolo (A. Chiò, G.R.), Adelis Foundation (V.E.D.), European Community's Health Seventh Framework Programme (FP7/2007-2013) under grant agreements 259867 (A. Chiò, M. Sendtner, C.D.), EuroMOTOR (M. Sendtner), German Federal Ministry of Education and Research (BMBF) (M. Sendtner), German Network for Motoneuron Disease (M. Sendtner, grant TP4) and NIH grant NS061867 (R.B.).
Author information
Authors and Affiliations
Consortia
Contributions
J.O.J., A.B., R.C., A.E.R., H.A.P., Y.A., G. Marangi, B.J.W., S.M., M. Shoai, A. Pittman, P.F., M.B.H., R.H.B., A. Pestronk and C.C.W. performed laboratory-based experiments and data analysis and revised the report; E.P.P., H.F., R.W.O., A.M., K.C.S., E.R., L.Z., V.E.D., G.B., G. Mora, A. Calvo, J.D.R., ITALSGEN, C.D., M. Sendtner, G.R., M. Sabatelli and A. Chiò collected clinical information and DNA samples and revised the report; J.R.G. and M.A.N. performed data analysis and revised the report; J.H., A.B.S., J.P.T., M.R.C. and R.B. supervised laboratory-based experiments and data analysis and revised the report; B.J.T. supervised the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
B.J.T., A.B.S. and J.H. have a patent pending on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9ORF72. J.D.R. has patents pending on antisense therapy for C9ORF72 and associated biomarkers, and is Director of the Packard Center for ALS Research at Johns Hopkins, which funded this study in part.
Additional information
Full lists of members and affiliations appear in the Supplementary Note.
Integrated supplementary information
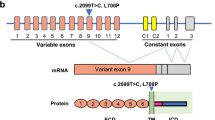
Supplementary Figure 1 Distribution of MATR3 mutations detected in familial ALS patients.
The upper panel shows the location of detected mutations and of the domains of MATR3 as determined by Hibino, Y., et al. Biochim. Biophys. Acta 1759, 195–207 (2006). Corresponding chromatograms showing mutant and wild-type alleles are as indicated, and conservation of amino acid residue across species is highlighted at the bottom (generated using the Clustal Omega online tool, www.ebi.ac.uk/Tools/msa/clustalo/).
Supplementary Figure 2 Lumbar spinal cord tissue immunostained for MATR3 and counterstained with hematoxylin.
(a) Control spinal cord exhibits MATR3 nuclear immunoreactivity in some motor neurons, with weak glial cell immunostaining. (b) Spinal cord from a subject with ALS exhibits strong nuclear immunoreactivity, with cytoplasmic immunoreactivity present in some motor neurons either diffusely or in cytoplasmic puncta. Strong glial immunostaining is also noted in samples from ALS patients. (c) Spinal cord from a patient with the Phe115Cys MATR3 mutation exhibits strong nuclear staining, as well as cytoplasmic staining in many cells. Images were taken at 20x magnification; insets show enlargements of the boxed regions. Scale bars represent 50 μm.
Supplementary Figure 3 MATR3-immunoreactive staining in spinal cord neurons of ALS patients.
(a & d) Control cases exhibit a nuclear staining pattern with staining not filling the entire nucleus. (b & e) ALS cases display stronger nuclear staining pattern with cytoplasmic staining present in some cells. Cytoplasmic staining is either diffuse across the entire cell or found in cytoplasmic puncta. E shows a MATR3-positive cytoplasmic inclusion, which are occasionally observed (this patient was known to carry a pathogenic C9ORF72 repeat expansion). (c & f) Patient carrying the pPhe115Cys MATR3 mutation shows strong nuclear staining and cytoplasmic staining in many cells. Immunohistochemistry was performed using the HPA036565 antibody (Sigma-Aldrich). Similar results were seen with a different anti-MATR3 antibody. MATR3 was not detected within cytoplasmic inclusions containing TDP-43 (data not shown). All images were taken at 40x magnification, and the scale bars represent 25μm.
Supplementary Figure 4 MATR3 and TDP-43 interaction is RNA dependent.
FLAG-MATR3 was expressed in 293FT cells, immunoprecipitated using anti-FLAG antibody followed by treatment with RNase A and probed with TDP-43 and DHX9 antibodies. Representative blots from two independent experiments are shown. Interaction of MATR3 and DHX9 is consistent with Salton, M. et al., PLoS One 6, e23882 (2011) showing that the interaction is RNA dependent, as is the interaction with TDP-43.
Supplementary Figure 5 Coimmunoprecipitation experiments using endogenous MATR3.
Endogenous MATR3 was immunoprecipitated from 293FT cells and probed with DHX9 and TDP-43 antibodies. Representative blots from two independent experiments are shown. These data show that MATR3, TDP-43 and DHX9 interact at the endogenous level.
Supplementary Figure 6 Immunofluorescence of skeletal muscle biopsies using anti-TDP-43 and anti-MATR3 antibodies.
Immunofluorescence of skeletal muscle biopsy from (a) a normal control, and (b) a patient carrying the p.Ser85Cys missense mutation in MATR3 using anti-TDP-43 and anti-MATR3 antibodies. In normal skeletal muscle, MATR3 and TDP-43 localize to nuclei including myonuclei. In the patient with the MATR3 mutation, there is decreased nuclear MATR3 immunoreactivity, whereas TDP-43 accumulates in the sarcoplasm and is restricted from the nucleus. In addition, MATR3 and TDP-43 co-aggregate in the sarcoplasm adjacent to myonuclei. Open arrow highlights a TDP-43 positive fiber (outlined in white). Closed arrows demonstrate MATR3 and TDP-43 co-localized in perinuclear inclusions. Insets are enlarged myonuclei and the scale bar is 50uM.
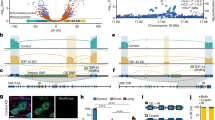
Supplementary Figure 8 Novel coding variants identified in the USALS#3 kindred by exome sequencing.
Graphical representation of autosomes showing genomic regions shared by the four affected individuals of the USALS#3 pedigree (blue lines). Whole genome data was generated using Infinium OmniExpress genotyping arrays (Illumina Inc.). LMNB1 and MATR3 variants are shown as red crosses located within a 17.4 Mb shared segment on chromosome 5q. No other novel, coding variants were shared across affected individuals of the USALS#3 family.
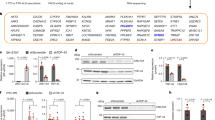
Supplementary Figure 9 Characterization of MATR3 antibodies.
The specificity of MATR3 antibodies Abcam 151714 (a) and Sigma HPA0036564 (b) were tested against purified MATR3 protein (Origene TP323258), lysates from HEK293FT cells treated with siRNA (untransfected control, non-targeting control, Cyclophilin B and MATR3), and cells overexpressing MATR3(WT)-GFP. Both antibodies were specific to MATR3 as indicated by the MATR3 siRNA sample showing a reduction in MATR3 protein level compared to untransfected, non-targeting and Cyclophilin B controls.
Supplementary Figure 10 Full-length blots.
Full-length blots for (a) Figure 3, (b) Supplementary Figure 3, and (c) Supplementary Figure 4.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Note (PDF 17073 kb)
Rights and permissions
About this article
Cite this article
Johnson, J., Pioro, E., Boehringer, A. et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci 17, 664–666 (2014). https://doi.org/10.1038/nn.3688
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.3688
This article is cited by
-
ASPSCR1-TFE3 reprograms transcription by organizing enhancer loops around hexameric VCP/p97
Nature Communications (2024)
-
Amyotrophic lateral sclerosis: translating genetic discoveries into therapies
Nature Reviews Genetics (2023)
-
Modelling amyotrophic lateral sclerosis in rodents
Nature Reviews Neuroscience (2022)
-
Phase separation of low-complexity domains in cellular function and disease
Experimental & Molecular Medicine (2022)
-
Ultrastructural and biochemical classification of pathogenic tau, α-synuclein and TDP-43
Acta Neuropathologica (2022)