Abstract
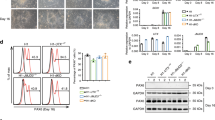
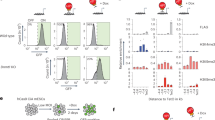
Signaling mediated by Notch receptors is crucial for the development of many organs and the maintenance of various stem cell populations. The activation of Notch signaling is first detectable by the expression of an effector gene, Hes5, in the neuroepithelium of mouse embryos at embryonic day (E) 8.0–8.5, and this activation is indispensable for the generation of neural stem cells. However, the molecular mechanism by which Hes5 expression is initiated in stem-producing cells remains unknown. We found that mammalian Gcm1 and Gcm2 (glial cells missing 1 and 2) are involved in the epigenetic regulation of Hes5 transcription by DNA demethylation independently of DNA replication. Loss of both Gcm genes and subsequent lack of Hes5 upregulation in the neuroepithelium of E7.5–8.5 Gcm1−/−; Gcm2−/− mice resulted in the impaired induction of neural stem cells. Our data suggest that Hes5 expression is serially activated first by Gcms and later by the canonical Notch pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Reynolds, B.A., Tetzlaff, W. & Weiss, S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J. Neurosci. 12, 4565–4574 (1992).
Hitoshi, S. et al. Primitive neural stem cells from the mammalian epiblast differentiate to definitive neural stem cells under the control of Notch signaling. Genes Dev. 18, 1806–1811 (2004).
Nakamura, Y. et al. The bHLH gene Hes1 as a repressor of the neuronal commitment of CNS stem cells. J. Neurosci. 20, 283–293 (2000).
Ohtsuka, T., Sakamoto, M., Guillemot, F. & Kageyama, R. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J. Biol. Chem. 276, 30467–30474 (2001).
Hitoshi, S. et al. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 16, 846–858 (2002).
Artavanis-Tsakonas, S., Rand, M.D. & Lake, R.J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
de la Pompa, J.L. et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development 124, 1139–1148 (1997).
Donoviel, D.B. et al. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13, 2801–2810 (1999).
Hosoya, T., Takizawa, K., Nitta, K. & Hotta, Y. glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell 82, 1025–1036 (1995).
Jones, B.W., Fetter, R.D., Tear, G. & Goodman, C.S. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell 82, 1013–1023 (1995).
Jones, B.W. Transcriptional control of glial cell development in Drosophila. Dev. Biol. 278, 265–273 (2005).
Akiyama, Y., Hosoya, T., Poole, A.M. & Hotta, Y. The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals. Proc. Natl. Acad. Sci. USA 93, 14912–14916 (1996).
Schreiber, J., Sock, E. & Wegner, M. The regulator of early gliogenesis glial cells missing is a transcription factor with a novel type of DNA-binding domain. Proc. Natl. Acad. Sci. USA 94, 4739–4744 (1997).
Egger, B. et al. Gliogenesis in Drosophila: genome-wide analysis of downstream genes of glial cells missing in the embryonic nervous system. Development 129, 3295–3309 (2002).
Kim, J. et al. Isolation and characterization of mammalian homologs of the Drosophila gene glial cells missing. Proc. Natl. Acad. Sci. USA 95, 12364–12369 (1998).
Günther, T. et al. Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature 406, 199–203 (2000).
Schreiber, J., Enderich, J. & Wegner, M. Structural requirement for DNA binding of GCM proteins. Nucleic Acids Res. 26, 2337–2343 (1998).
Schreiber, J. et al. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol. Cell. Biol. 20, 2466–2474 (2000).
Hatakeyama, J. et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131, 5539–5550 (2004).
Martens, D.J., Tropepe, V. & van der Kooy, D. Separate proliferation kinetics of fibroblast growth factor–responsive and epidermal growth factor–responsive neural stem cells within the embryonic forebrain germinal zone. J. Neurosci. 20, 1085–1095 (2000).
Wolffe, A.P., Jones, P.L. & Wade, P.A. DNA demethylation. Proc. Natl. Acad. Sci. USA 96, 5894–5896 (1999).
Zhu, J.-K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43, 143–166 (2009).
Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 (2007).
Waga, S. & Stillman, B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751 (1998).
Bruniquel, D. & Schwartz, R.H. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat. Immunol. 4, 235–240 (2003).
Ma, D.K. et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077 (2009).
Kim, M.-S. et al. DNA demethylation in hormone-induced transcriptional derepression. Nature 461, 1007–1012 (2009).
Okada, Y., Yamagata, K., Hong, K., Wakayama, T. & Zhang, Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature 463, 554–558 (2010).
Rai, K. et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell 135, 1201–1212 (2008).
Kimura, C. et al. Visceral endoderm mediates forebrain development by suppressing posteriorizing signals. Dev. Biol. 225, 304–321 (2000).
Zhu, B. et al. 5-Methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc. Natl. Acad. Sci. USA 97, 5135–5139 (2000).
Cau, E., Gradwohl, G., Casarosa, S., Kageyama, R. & Guillemot, F. Hes genes regulates sequential stages of neurogenesis in the olfactory epithelium. Development 127, 2323–2332 (2000).
Kageyama, R., Ohtsuka, T. & Kobayashi, T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251 (2007).
Oka, C. et al. Disruption of the mouse RBP-Jκ gene results in early embryonic death. Development 121, 3291–3301 (1995).
Kageyama, R., Ohtsuka, T., Shimojo, H. & Imayoshi, I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 11, 1247–1251 (2008).
Bettenhausen, B., Hrabĕ de Angelis, M., Simon, D., Guénet, J.L. & Gossler, A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development 121, 2407–2418 (1995).
Hatakeyama, J. & Kageyama, R. Notch1 expression is spatiotemporally correlated with neurogenesis and negatively regulated by Notch1-independent Hes genes in the developing nervous system. Cereb. Cortex 16 (suppl. 1), i132–i137 (2006).
Hirata, H. et al. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843 (2002).
Shimojo, H., Ohtsuka, T. & Kageyama, R. Oscillations in Notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64 (2008).
Nakahira, E., Kagawa, T., Shimizu, T., Goulding, M.D. & Ikenaka, K. Direct evidence that ventral forebrain cells migrate to the cortex and contribute to the generation of cortical myelinating oligodendrocytes. Dev. Biol. 291, 123–131 (2006).
Iwasaki, Y. et al. The potential to induce glial differentiation is conserved between Drosophila and mammalian glial cells missing genes. Development 130, 6027–6035 (2003).
Acknowledgements
We thank R. Kageyama, J.S. Nye, T. Miyazaki and T. Honjo for plasmids, T.-e.U and Y. Imai for MBD4 mutant mice, and R. Kageyama, D. van der Kooy and K. Nakashima for comments. This work was supported by a Grant-in-Aid for Exploratory Research (19650096) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (K.I.S.H.).
Author information
Authors and Affiliations
Contributions
S.H. designed and carried out the experiments, analyzed the data and wrote the paper. Y.I., A.K., S.J., K.F.T. and T.H. generated Gcm mutant mice and analyzed the phenotypes. T.K. and S.K. carried out the experiments related to MBD4 knockout mice. Y.H. and K.I. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1–9 and Supplementary Table 1 (PDF 2603 kb)
Rights and permissions
About this article
Cite this article
Hitoshi, S., Ishino, Y., Kumar, A. et al. Mammalian Gcm genes induce Hes5 expression by active DNA demethylation and induce neural stem cells. Nat Neurosci 14, 957–964 (2011). https://doi.org/10.1038/nn.2875
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2875
This article is cited by
-
Analysis of cnidarian Gcm suggests a neuronal origin of glial EAAT1 function
Scientific Reports (2023)
-
Lineage of drug discovery research on fluorinated pyrimidines: chronicle of the achievements accomplished by Professor Setsuro Fujii
International Journal of Clinical Oncology (2023)
-
The hypoparathyroidism-associated mutation in Drosophila Gcm compromises protein stability and glial cell formation
Scientific Reports (2017)
-
Ten-Eleven Translocation 1 and 2 Confer Overlapping Transcriptional Programs for the Proliferation of Cultured Adult Neural Stem Cells
Cellular and Molecular Neurobiology (2017)
-
An evolutionary conserved interaction between the Gcm transcription factor and the SF1 nuclear receptor in the female reproductive system
Scientific Reports (2016)