Abstract
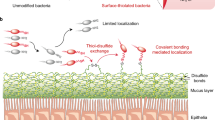
The immunomodulatory surface molecules of commensal and pathogenic bacteria are critical to microorganisms' survival and the host's response1,2. Recent studies have highlighted the unique and important responses elicited by commensal-derived surface macromolecules3–5. However, the technology available to track these molecules in host cells and tissues remains primitive. We report, here, an interdisciplinary approach that uses metabolic labelling combined with bioorthogonal click chemistry (that is, reactions performed in living organisms)6 to specifically tag up to three prominent surface immunomodulatory macromolecules—peptidoglycan, lipopolysaccharide and capsular polysaccharide—either simultaneously or individually in live anaerobic commensal bacteria. Importantly, the peptidoglycan labelling enables, for the first time, the specific labelling of live endogenous, anaerobic bacteria within the mammalian host. This approach has allowed us to image and track the path of labelled surface molecules from live, luminal bacteria into specific intestinal immune cells in the living murine host during health and disease. The chemical labelling of three specific macromolecules within a live organism offers the potential for in-depth visualization of host–pathogen interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fischbach, M. A. & Segre, J. A. Signaling in host-associated microbial communities. Cell 164, 1288–1300 (2016).
Sommer, F. & Backhed, F. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238 (2013).
Ayres, J. S. Cooperative microbial tolerance behaviors in host–microbiota mutualism. Cell 165, 1323–1331 (2016).
Lebeer, S., Vanderleyden, J. & De Keersmaecker, S. C. J. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8, 171–184 (2010).
Rooks, M. G. & Garrett, W. S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352 (2016).
Sletten, E. M. & Bertozzi, C. R. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 48, 6974–6998 (2009).
Earle, K. A. et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 18, 478–488 (2015).
Welch, J. L. M., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E. & Borisy, G. G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl Acad. Sci. USA 113, E791–E800 (2016).
Moter, A. & Göbel, U. B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 41, 85–112 (2000).
Siegrist, M. S., Swarts, B. M., Fox, D. M., Lim, S. A. & Bertozzi, C. R. Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiol. Rev. 39, 184–202 (2015).
Kocaoglu, O. & Carlson, E. E. Progress and prospects for small-molecule probes of bacterial imaging. Nat. Chem. Biol. 12, 472–478 (2016).
Geva-Zatorsky, N. et al. In vivo imaging and tracking of host–microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat. Med. 21, 1091–1100 (2015).
Boyce, M. & Bertozzi, C. R. Bringing chemistry to life. Nat. Methods 8, 638–642 (2011).
Thaiss, C. A., Levy, M., Suez, J. & Elinav, E. The interplay between the innate immune system and the microbiota. Curr. Opin. Immunol. 26, 41–48 (2014).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 (2001).
Kuru, E. et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew. Chem. Int. Ed. 51, 12519–12523 (2012).
Hsu, Y.-P., Meng, X. & VanNieuwenhze, M. S. in Methods in Microbiology Vol. 43 (eds Jensen, G. J. & Harwood, C. ) 3–48 (Academic, 2016).
Mowat, A. M. & Agace, W. W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 14, 667–685 (2014).
Helmchen, F. & Denk, W. Deep tissue two-photon microscopy. Nat. Methods 2, 932–940 (2005).
Farache, J. et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 38, 581–595 (2013).
Mazzini, E., Massimiliano, L., Penna, G. & Rescigno, M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1+ macrophages to CD103+ dendritic cells. Immunity 40, 248–261 (2014).
Reboldi, A. et al. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer's patches. Science 352, aaf4822 (2016).
Diehl, G. E. et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 494, 116–120 (2013).
Sánchez de Medina, F., Romero-Calvo, I., Mascaraque, C. & Martínez-Augustin, O. Intestinal inflammation and mucosal barrier function. Inflamm. Bowel Dis. 20, 2394–2404 (2014).
Sartor, R. B. Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594 (2008).
Dumont, A., Malleron, A., Awwad, M., Dukan, S. & Vauzeilles, B. Click-mediated labeling of bacterial membranes through metabolic modification of the lipopolysaccharide inner core. Angew. Chem. Int. Ed. 51, 3143–3146 (2012).
Kumada, H., Haishima, Y., Kondo, S., Umemoto, T. & Hisatsune, K. Occurrence of 2-keto-3-deoxyoctonate (KDO) and KDO phosphate in lipopolysaccharides of Bacteriodes species. Curr. Microbiol. 26, 239–244 (1993).
Patterson, D. M., Jones, K. A. & Prescher, J. A. Improved cyclopropene reporters for probing protein glycosylation. Mol. Biosyst. 10, 1693–1697 (2014).
Patterson, D. M., Nazarova, L. A., Xie, B., Kamber, D. N. & Prescher, J. A. Functionalized cyclopropenes as bioorthogonal chemical reporters. J. Am. Chem. Soc. 134, 18638–18643 (2012).
Elson, C. O. et al. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol. Rev. 206, 260–276 (2005).
Wirtz, S., Neufert, C., Weigmann, B. & Neurath, M. F. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2, 541–546 (2007).
Couter, C. J. & Surana, N. K. Isolation and flow cytometric characterization of murine small intestinal lymphocytes. J. Vis. Exp. 111, e54114 (2016).
Millet, Y. A. et al. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 10, e1004405 (2014).
Thévenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (1998).
Saalfeld, S., Fetter, R., Cardona, A. & Tomancak, P. Elastic volume reconstruction from series of ultra-thin microscopy sections. Nat. Methods 9, 717–720 (2012).
Acknowledgements
The authors acknowledge the Harvard Medical School Center for Immune Imaging for providing instrumentation and aid for two-photon microscopy. The authors thank N. Geva-Zatorsky and F. Gazzaniga for materials, expertise and discussion, D. Erturk-Hasdemir and N. Okan for aid in cell culture and isolation of bone marrow macrophages, and C. Hudak for discussion and manuscript critique. This work was funded by a grant from the US Department of Defense (W81XWH-15-1-0368) and was supported in part by the US National Institutes of Health (grants PO1 AI1112521, RO1 AI111595 (to U.H.v.A.) and 5T32 HL066987 (to D.A.)). Additional support to U.H.v.A. was provided by the Ragon Institute at MGH, MIT and Harvard. J.E.H. was supported by the Cancer Research Institute Irvington Fellowship Program.
Author information
Authors and Affiliations
Contributions
J.E.H. designed the experiments, analysed the data and wrote the manuscript with help from D.A. and A.S. D.A. provided expertise in two-photon intravital microscopy. D.L.K. supervised the study, edited the manuscript and provided helpful comments, with assistance from U.H.v.A.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–13, Supplementary Notes and Supplementary References. (PDF 18026 kb)
Supplementary Video 1
Intravital two-photon microscopy of the commensal microflora in the mouse colon. (MOV 12295 kb)
Supplementary Video 2
Intravital two-photon microscopy of the commensal microflora in the mouse colon. (MOV 11091 kb)
Supplementary Video 3
Intravital two-photon microscopy of the commensal microbe B. vulgatus in the mouse small intestine. (MOV 4731 kb)
Supplementary Video 4
Intravital two-photon microscopy of the commensal microbe B. vulgatus in the mouse small intestine. (MOV 6610 kb)
Rights and permissions
About this article
Cite this article
Hudak, J., Alvarez, D., Skelly, A. et al. Illuminating vital surface molecules of symbionts in health and disease. Nat Microbiol 2, 17099 (2017). https://doi.org/10.1038/nmicrobiol.2017.99
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2017.99
This article is cited by
-
Omics for deciphering oral microecology
International Journal of Oral Science (2024)
-
Engineered live bacteria as disease detection and diagnosis tools
Journal of Biological Engineering (2023)
-
Combinatorial fluorescent labeling of live anaerobic bacteria via the incorporation of azide-modified sugars into newly synthesized macromolecules
Nature Protocols (2023)
-
Extracellular vesicles of the Gram-positive gut symbiont Bifidobacterium longum induce immune-modulatory, anti-inflammatory effects
npj Biofilms and Microbiomes (2023)
-
Selective inactivation of Gram-positive bacteria in vitro and in vivo through metabolic labelling
Science China Materials (2022)