Abstract
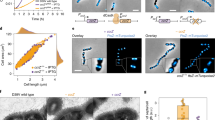
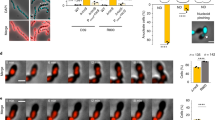
Cell division must be coordinated with chromosome replication and segregation to ensure the faithful transmission of genetic information during proliferation. In most bacteria, assembly of the division apparatus, the divisome, starts with the polymerization of a tubulin homologue, FtsZ, into a ring-like structure at mid-cell, the Z-ring1. It typically occurs at half of the cell cycle when most of the replication and segregation cycle of the unique chromosome they generally harbour is achieved2. The chromosome itself participates in the regulation of cell division, at least in part because it serves as a scaffold to position FtsZ polymerization antagonists3. However, about 10% of bacteria have more than one chromosome4, which raises questions about the way they license cell division3. For instance, the genome of Vibrio cholerae, the agent of cholera, is divided between a 3 Mbp replicon that originates from the chromosome of its mono-chromosomal ancestor, Chr1, and a 1 Mbp plasmid-derived replicon, Chr2 (ref. 5). Here, we show that Chr2 harbours binding motifs for an inhibitor of Z-ring formation, which helps accurately position the V. cholerae divisome at mid-cell and postpones its assembly to the very end of the cell cycle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bi, E. F. & Lutkenhaus, J. FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 (1991).
den Blaauwen, T. Prokaryotic cell division: flexible and diverse. Curr. Opin. Microbiol. 16, 738–744 (2013).
Adams, D. W., Wu, L. J. & Errington, J. Cell cycle regulation by the bacterial nucleoid. Curr. Opin. Microbiol. 22, 94–101 (2014).
Val, M.-E., Soler-Bistué, A., Bland, M. J. & Mazel, D. Management of multipartite genomes: the Vibrio cholerae model. Curr. Opin. Microbiol. 22, 120–126 (2014).
Heidelberg, J. F. et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483 (2000).
Srivastava, P., Fekete, R. A. & Chattoraj, D. K. Segregation of the replication terminus of the two Vibrio cholerae chromosomes. J. Bacteriol. 188, 1060–1070 (2006).
Val, M.-E. et al. FtsK-dependent dimer resolution on multiple chromosomes in the pathogen Vibrio cholerae. PLoS Genet. 4, e1000201 (2008).
Demarre, G., Galli, E. & Barre, F.-X. The ftsK family of DNA pumps. Adv. Exp. Med. Biol. 767, 245–262 (2013).
De Boer, P. A., Crossley, R. E. & Rothfield, L. I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56, 641–649 (1989).
Ortiz, C., Natale, P., Cueto, L. & Vicente, M. The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol. Rev. 40, 57–67 (2016).
Lutkenhaus, J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76, 539–562 (2007).
David, A. et al. The two cis-acting sites, parS1 and oriC1, contribute to the longitudinal organisation of Vibrio cholerae chromosome I. PLoS Genet. 10, e1004448 (2014).
Fogel, M. A. & Waldor, M. K. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 20, 3269–3282 (2006).
Demarre, G. et al. Differential management of the replication terminus regions of the two Vibrio cholerae chromosomes during cell division. PLoS Genet. 10, e1004557 (2014).
Mercier, R. et al. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135, 475–485 (2008).
Woldringh, C. L., Mulder, E., Huls, P. G. & Vischer, N. Toporegulation of bacterial division according to the nucleoid occlusion model. Res. Microbiol. 142, 309–320 (1991).
Bernhardt, T. G. & de Boer, P. A. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18, 555–564 (2005).
Val, M.-E., Skovgaard, O., Ducos-Galand, M., Bland, M. J. & Mazel, D. Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet. 8, e1002472 (2012).
Cambridge, J., Blinkova, A., Magnan, D., Bates, D. & Walker, J. R. A replication-inhibited unsegregated nucleoid at mid-cell blocks Z-ring formation and cell division independently of SOS and the SlmA nucleoid occlusion protein in Escherichia coli. J. Bacteriol. 196, 36–49 (2014).
Bailey, M. W., Bisicchia, P., Warren, B. T., Sherratt, D. J. & Männik, J. Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet. 10, e1004504 (2014).
Rasmussen, T., Jensen, R. B. & Skovgaard, O. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 26, 3124–3131 (2007).
Sliusarenko, O., Heinritz, J., Emonet, T. & Jacobs-Wagner, C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80, 612–627 (2011).
Acknowledgements
The authors thank M.-E. Val and D. Mazel for the gift of strain MCH1 and N. Dubarry and the other team members for insightful comments. The authors acknowledge financial support from the European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013 grant agreement no. 281590), the Fondation Bettencourt Schueller (2012 Coup d'Elan award) and the ‘Lidex-Biologie Intégrative des Génomes’ project of the Paris-Saclay IDEX (ANR-11-IDEX-0003-02). The light microscopy facility of Imagerie-Gif is a member of the Infrastructures en Biologie Santé et Agronomie (IBiSA), and is supported by the French national Research Agency under Investments for the Future programmes ‘France-BioImaging’ and the Labex ‘Saclay Plant Science’ (ANR-10-INSB-04-01 and ANR-11-IDEX-0003-02, respectively).
Author information
Authors and Affiliations
Contributions
E.G., M.P., Y.Y. and F.X.B. conceived and designed the experiments. E.G., M.P., R.L.B. and E.P. performed the experiments. E.G., M.P., Y.Y., J.-M.D. and F.X.B. analysed the data. E.G., J.-M.D. and L.M. contributed reagents/materials/analysis tools. E.G., M.P., Y.Y. and F.X.B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Figures 1-10, Supplementary Tables 1-5 and Supplementary References. (PDF 7635 kb)
Supplementary Video 1
3D FtsZ-mEos2 structures throughout the V. cholerae cell cycle observed using PALM. Cells grown in M9 minimal medium supplemented with 0.2% fructose and 1 µg ml−1 thiamine. (MP4 6440 kb)
Supplementary Video 2
Time-lapse analysis of MinD-YGFP localization in V. cholerae cells. One frame was taken every 20 seconds. Cells grown in M9 minimal medium supplemented with 0.2% fructose and 1 µg ml−1 thiamine (AVI 190 kb)
Supplementary Video 3
Time-lapse analysis of MinD-YGFP and FtsZ-RFPT localization in V. cholerae cells. One frame was taken every 20 seconds. Cells grown in M9 minimal medium supplemented with 0.2% fructose and 1 µg ml−1 thiamine. (AVI 207 kb)
Supplementary Video 4
Mini-cell formation in MCH1. One frame was taken every 3 minutes. Cells were grown in LB (AVI 115 kb)
Supplementary Video 5
Formation of an anucleate cell after a highly asymmetric division in MCH1. Genomic DNA is labelled with a HUα-RFPT fusion. One frame was taken every 3 minutes. Cells grown in M9 minimal medium supplemented with 0.2% fructose and 1 µg ml−1 thiamine. (AVI 55 kb)
Rights and permissions
About this article
Cite this article
Galli, E., Poidevin, M., Le Bars, R. et al. Cell division licensing in the multi-chromosomal Vibrio cholerae bacterium. Nat Microbiol 1, 16094 (2016). https://doi.org/10.1038/nmicrobiol.2016.94
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.94
This article is cited by
-
Replication termination without a replication fork trap
Scientific Reports (2019)
-
The E. coli MinCDE system in the regulation of protein patterns and gradients
Cellular and Molecular Life Sciences (2019)
-
Late assembly of the Vibrio cholerae cell division machinery postpones septation to the last 10% of the cell cycle
Scientific Reports (2017)