Abstract
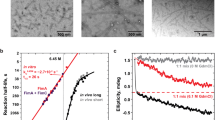
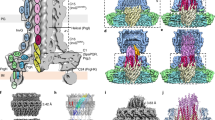
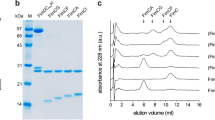
Outer membrane proteins are essential for Gram-negative bacteria to rapidly adapt to changes in their environment. Intricate remodelling of the outer membrane proteome is critical for bacterial pathogens to survive environmental changes, such as entry into host tissues1–3. Fimbriae (also known as pili) are appendages that extend up to 2 μm beyond the cell surface to function in adhesion for bacterial pathogens, and are critical for virulence. The best-studied examples of fimbriae are the type 1 and P fimbriae of uropathogenic Escherichia coli, the major causative agent of urinary tract infections in humans. Fimbriae share a common mode of biogenesis, orchestrated by a molecular assembly platform called ‘the usher’ located in the outer membrane. Although the mechanism of pilus biogenesis is well characterized, how the usher itself is assembled at the outer membrane is unclear. Here, we report that a rapid response in usher assembly is crucially dependent on the translocation assembly module. We assayed the assembly reaction for a range of ushers and provide mechanistic insight into the β-barrel assembly pathway that enables the rapid deployment of bacterial fimbriae.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vogel, J. & Papenfort, K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9, 605–611 (2006).
Clegg, S., Wilson, J. & Johnson, J. More than one way to control hair growth: regulatory mechanisms in enterobacteria that affect fimbriae assembled by the chaperone/usher pathway. J. Bacteriol. 193, 2081–2088 (2011).
Dalebroux, Z. D. et al. Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17, 441–451 (2015).
McMorran, L. M., Brockwell, D. J. & Radford, S. E. Mechanistic studies of the biogenesis and folding of outer membrane proteins in vitro and in vivo: what have we learned to date? Arch. Biochem. Biophys. 564, 265–280 (2014).
Selkrig, J., Leyton, D. L., Webb, C. T. & Lithgow, T. Assembly of β-barrel proteins into bacterial outer membranes. Biochim. Biophys. Acta 1843, 1542–1550 (2014).
Fleming, K. G. A combined kinetic push and thermodynamic pull as driving forces for outer membrane protein sorting and folding in bacteria. Phil. Trans. R. Soc. Lond. B 370, 20150026 (2015).
O'Neil, P. K., Rollauer, S. E., Noinaj, N. & Buchanan, S. K. Fitting the pieces of the β-barrel assembly machinery complex. Biochemistry 54, 6303–6311 (2015).
Heinz, E., Selkrig, J., Belousoff, M. J. & Lithgow, T. Evolution of the translocation and assembly module (TAM). Genome Biol. Evol. 7, 1628–1643 (2015).
Noinaj, N. et al. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 (2013).
Noinaj, N., Kuszak, A. J., Balusek, C., Gumbart, J. C. & Buchanan, S. K. Lateral opening and exit pore formation are required for BamA function. Structure 22, 1055–1062 (2014).
Selkrig, J. et al. Discovery of an archetypal protein transport system in bacterial outer membranes. Nature Struct. Mol. Biol. 19, 506–510 (2012).
Gruss, F. et al. The structural basis of autotransporter translocation by TamA. Nature Struct. Mol. Biol. 20, 1318–1320 (2013).
Shen, H. H. et al. Reconstitution of a nanomachine driving the assembly of proteins into bacterial outer membranes. Nature Commun. 5, 5078 (2014).
Selkrig, J. et al. Conserved features in TamA enable interaction with TamB to drive the activity of the translocation and assembly module. Sci. Rep. 5, 12905 (2015).
Phan, G. et al. Crystal structure of the FimD usher bound to its cognate FimC–FimH substrate. Nature 474, 49–53 (2011).
Geibel, S., Procko, E., Hultgren, S. J., Baker, D. & Waksman, G. Structural and energetic basis of folded-protein transport by the FimD usher. Nature 496, 243–246 (2013).
Stenberg, F., von Heijne, G. & Daley, D. O. Assembly of the cytochrome bo 3 complex. J. Mol. Biol. 371, 765–773 (2007).
Noinaj, N., Rollauer, S. E. & Buchanan, S. K. The β-barrel membrane protein insertase machinery from Gram-negative bacteria. Curr. Opin. Struct. Biol. 31, 35–42 (2015).
De Cock, H., van Blokland, S. & Tommassen, J. In vitro insertion and assembly of outer membrane protein PhoE of Escherichia coli K-12 into the outer membrane. Role of Triton X-100. J. Biol. Chem. 271, 12885–12890 (1996).
Werner, J. & Misra, R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 57, 1450–1459 (2005).
Korea, C. G., Ghigo, J. M. & Beloin, C. The sweet connection: Solving the riddle of multiple sugar-binding fimbrial adhesins in Escherichia coli: multiple E. coli fimbriae form a versatile arsenal of sugar-binding lectins potentially involved in surface-colonisation and tissue tropism. BioEssays: News Rev. Mol. Cell. Dev. Biol. 33, 300–311 (2011).
Busch, A. & Waksman, G. Chaperone-usher pathways: diversity and pilus assembly mechanism. Phil. Trans. R. Soc. Lond. B 367, 1112–1122 (2012).
Wu, X. R., Sun, T. T. & Medina, J. J. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc. Natl Acad. Sci. USA 93, 9630–9635 (1996).
Schembri, M. A., Sokurenko, E. V. & Klemm, P. Functional flexibility of the FimH adhesin: insights from a random mutant library. Infect. Immun. 68, 2638–2646 (2000).
Nilsson, L. M., Thomas, W. E., Trintchina, E., Vogel, V. & Sokurenko, E. V. Catch bond-mediated adhesion without a shear threshold: trimannose versus monomannose interactions with the FimH adhesin of Escherichia coli. J. Biol. Chem. 281, 16656–16663 (2006).
Nuccio, S. P. & Baumler, A. J. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71, 551–575 (2007).
Kuehn, M. J., Heuser, J., Normark, S. & Hultgren, S. J. P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature 356, 252–255 (1992).
Iguchi, A. et al. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191, 347–354 (2009).
Korea, C. G., Badouraly, R., Prevost, M. C., Ghigo, J. M. & Beloin, C. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ. Microbiol. 12, 1957–1977 (2010).
Wurpel, D. J., Beatson, S. A., Totsika, M., Petty, N. K. & Schembri, M. A. Chaperone-usher fimbriae of Escherichia coli. PLoS ONE 8, e52835 (2013).
Remaut, H. et al. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133, 640–652 (2008).
Stærk, K., Khandige, S., Kolmos, H. J., Møller-Jensen, J. & Andersen, T. E. Uropathogenic Escherichia coli express type 1 fimbriae only in surface adherent populations under physiological growth conditions. J. Infect. Dis. 213, 386–394 (2016).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Sambrook, J. & Russell, D. Molecular Cloning: A Laboratory Manual 3rd edn, Vol. 3, A2.2 (Cold Spring Harbor Laboratory Press, 2001).
Phu, L. et al. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell. Proteom. 10, M110.003756 (2011).
Wenger, C. D. & Coon, J. J. A proteomics search algorithm specifically designed for high-resolution tandem mass spectra. J. Proteome Res. 12, 1377–1386 (2013).
Schilling, B. et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol. Cell Proteom. 11, 202–214 (2012).
Wright, K. J., Seed, P. C. & Hultgren, S. J. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 9, 2230–2241 (2007).
Acknowledgements
The authors thank R. Goode and O. Kleinfeld for proteomic analysis and R. Bamert, R. Dunstan, R. Grinter and E. Heinz for comments on the manuscript. The authors also thank S. Hultgren for providing strain UTI89-Ptet-fim. This work was supported by an NHMRC Program Grant (606788, to T.L. and R.A.S.) and an NHMRC Project grant (APP1042651, to M.A.S.). T.L. is an ARC Australian Laureate Fellow, MA.S. is an NHMRC Senior Research Fellow, H.H.S. is an ARC Super Science Fellow, M.J.B. is an NHMRC Biomedical Fellow, and I.D.H. is an ARC Laureate Postdoctoral Fellow.
Author information
Authors and Affiliations
Contributions
C.S., M.J.B., I.D.H., K.M.P., M.-D.P. and A.W.L. designed and carried out the analyses. C.S., M.J.B., I.D.H., J.L., M.A.S., H.H.S. and K.L.T. provided expertise for the analyses. M.A.S., M.J.B., I.D.H., J.L., R.A.S., G.W. and T.L. supervised experimental work and evaluated data. M.A.S., R.A.S., G.W. and T.L. evaluated the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Figures 1–9 (PDF 1667 kb)
Rights and permissions
About this article
Cite this article
Stubenrauch, C., Belousoff, M., Hay, I. et al. Effective assembly of fimbriae in Escherichia coli depends on the translocation assembly module nanomachine. Nat Microbiol 1, 16064 (2016). https://doi.org/10.1038/nmicrobiol.2016.64
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.64
This article is cited by
-
Surveying membrane landscapes: a new look at the bacterial cell surface
Nature Reviews Microbiology (2023)
-
Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA β-barrel
Nature Communications (2019)
-
The Rich Tapestry of Bacterial Protein Translocation Systems
The Protein Journal (2019)