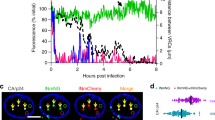
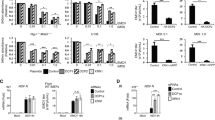
Abstract
The nucleus is highly compartmentalized yet dynamic. Subnuclear functions are regulated by controlling the subnuclear localization of the nuclear proteins. Influenza viral ribonucleoprotein (vRNP) is replicated in the nucleus and then exported to the cytoplasm. However, the precise subnuclear localization and transport of vRNPs remain unclear. Here, we show that CLUH, a host protein whose cellular function is not well established, plays a key role in the subnuclear transport of vRNP. Viral PB2 and M1 induced CLUH translocation to the nucleoplasm and SC35-positive speckles, respectively, even though CLUH is usually cytoplasmic. CLUH depletion inhibited the translocation of M1 to SC35-positive speckles, but did not interfere with PB2 localization to the nucleoplasm and disrupted the subnuclear transport of vRNP, abolishing vRNP nuclear export without affecting viral RNA or protein expression. Our findings suggest that CLUH plays a role in the subnuclear transport of progeny vRNP.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lamond, A. I. & Sleeman, J. E. Nuclear substructure and dynamics. Curr. Biol. 13, R825–R828 (2003).
Mao, Y. S., Zhang, B. & Spector, D. L. Biogenesis and function of nuclear bodies. Trends Genet. 27, 295–306 (2011).
Nickerson, J. A., Krockmalnic, G., Wan, K. M., Turner, C. D. & Penman, S. A normally masked nuclear matrix antigen that appears at mitosis on cytoskeleton filaments adjoining chromosomes, centrioles, and midbodies. J. Cell Biol. 116, 977–987 (1992).
Pederson, T. Half a century of ‘the nuclear matrix’. Mol. Biol. Cell 11, 799–805 (2000).
Chase, G. P. et al. Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting. PLoS Pathogens 7, e1002187 (2011).
Oh, Y. S. et al. SMARCA3, a chromatin-remodeling factor, is required for p11-dependent antidepressant action. Cell 152, 831–843 (2013).
Engelke, R. et al. The quantitative nuclear matrix proteome as a biochemical snapshot of nuclear organization. J. Proteome Res. 13, 3940–3956 (2014).
Saitoh, N. et al. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 15, 3876–3890 (2004).
Razin, S. V., Borunova, V. V., Iarovaia, O. V. & Vassetzky, Y. S. Nuclear matrix and structural and functional compartmentalization of the eucaryotic cell nucleus. Biochemistry 79, 608–618 (2014).
Eisfeld, A. J., Neumann, G. & Kawaoka, Y. At the centre: influenza A virus ribonucleoproteins. Nature Rev. Microbiol. 13, 28–41 (2015).
Amorim, M. J. & Digard, P. Influenza A virus and the cell nucleus. Vaccine 24, 6651–6655 (2006).
Josset, L., Frobert, E. & Rosa-Calatrava, M. Influenza A replication and host nuclear compartments: many changes and many questions. J. Clin. Virol. 43, 381–390 (2008).
Hutchinson, E. C. & Fodor, E. Transport of the influenza virus genome from nucleus to nucleus. Viruses 5, 2424–2446 (2013).
Watanabe, T., Watanabe, S. & Kawaoka, Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7, 427–439 (2010).
Martin, K. & Helenius, A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67, 117–130 (1991).
Neumann, G., Hughes, M. T. & Kawaoka, Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19, 6751–6758 (2000).
Elton, D. et al. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75, 408–419 (2001).
Boulo, S., Akarsu, H., Ruigrok, R. W. & Baudin, F. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 124, 12–21 (2007).
Paterson, D. & Fodor, E. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathogens 8, e1003019 (2012).
Eisfeld, A. J., Kawakami, E., Watanabe, T., Neumann, G. & Kawaoka, Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 85, 6117–6126 (2011).
Amorim, M. J. et al. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J. Virol. 85, 4143–4156 (2011).
Lopez-Turiso, J. A., Martinez, C., Tanaka, T. & Ortin, J. The synthesis of influenza virus negative-strand RNA takes place in insoluble complexes present in the nuclear matrix fraction. Virus Res. 16, 325–337 (1990).
Takizawa, N., Watanabe, K., Nouno, K., Kobayashi, N. & Nagata, K. Association of functional influenza viral proteins and RNAs with nuclear chromatin and sub-chromatin structure. Microbes Infect. 8, 823–833 (2006).
Engelhardt, O. G., Smith, M. & Fodor, E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 79, 5812–5818 (2005).
Ma, K., Roy, A. M. & Whittaker, G. R. Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology 282, 215–220 (2001).
Zhirnov, O. P. & Klenk, H. D. Histones as a target for influenza virus matrix protein M1. Virology 235, 302–310 (1997).
Garcia-Robles, I., Akarsu, H., Muller, C. W., Ruigrok, R. W. & Baudin, F. Interaction of influenza virus proteins with nucleosomes. Virology 332, 329–336 (2005).
Compans, R. W. & Dimmock, N. J. An electron microscopic study of single-cycle infection of chick embryo fibroblasts by influenza virus. Virology 39, 499–515 (1969).
Anisimova, E., Ghendon, Y. & Markushin, S. Ultrastructural changes in cells induced by temperature-sensitive mutants of fowl plague virus at permissive and non-permissive temperature. J. Gen. Virol. 47, 11–18 (1980).
Fortes, P., Lamond, A. I. & Ortin, J. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J. Gen. Virol. 76(Pt 4), 1001–1007 (1995).
Brunotte, L. et al. The nuclear export protein of H5N1 influenza A viruses recruits Matrix 1 (M1) protein to the viral ribonucleoprotein to mediate nuclear export. J. Biol. Chem. 289, 20067–20077 (2014).
Cox, R. T. & Spradling, A. C. Clueless, a conserved Drosophila gene required for mitochondrial subcellular localization, interacts genetically with parkin. Dis. Model Mech. 2, 490–499 (2009).
Gao, J. et al. CLUH regulates mitochondrial biogenesis by binding mRNAs of nuclear-encoded mitochondrial proteins. J. Cell Biol. 207, 213–223 (2014).
Sen, A., Damm, V. T. & Cox, R. T. Drosophila clueless is highly expressed in larval neuroblasts, affects mitochondrial localization and suppresses mitochondrial oxidative damage. PLoS ONE 8, e54283 (2013).
Watanabe, T. et al. Influenza virus–host interactome screen as a platform for antiviral drug development. Cell Host Microbe 16, 795–805 (2014).
Huang, S. et al. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J. Virol. 87, 767–778 (2013).
Iwatsuki-Horimoto, K., Horimoto, T., Fujii, Y. & Kawaoka, Y. Generation of influenza A virus NS2 (NEP) mutants with an altered nuclear export signal sequence. J. Virol. 78, 10149–10155 (2004).
Kawakami, E. et al. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J. Virol. Methods 173, 1–6 (2011).
Lamond, A. I. & Spector, D. L. Nuclear speckles: a model for nuclear organelles. Nature Rev. Mol. Cell Biol. 4, 605–612 (2003).
Spector, D. L. & Lamond, A. I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3, a000646 (2011).
Hu, Q. et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc. Natl Acad. Sci. USA 105, 19199–19204 (2008).
Brown, J. M. et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 182, 1083–1097 (2008).
Shopland, L. S., Johnson, C. V., Byron, M., McNeil, J. & Lawrence, J. B. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J. Cell Biol. 162, 981–990 (2003).
Kawaguchi, A., Matsumoto, K. & Nagata, K. YB-1 functions as a porter to lead influenza virus ribonucleoprotein complexes to microtubules. J. Virol. 86, 11086–11095 (2012).
Momose, F. et al. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277, 45306–45314 (2002).
Kawaguchi, A., Asaka, M. N., Matsumoto, K. & Nagata, K. Centrosome maturation requires YB-1 to regulate dynamic instability of microtubules for nucleus reassembly. Sci. Rep. 5, 8768 (2015).
Naito, T., Momose, F., Kawaguchi, A. & Nagata, K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 81, 1339–1349 (2007).
Morita, S., Kojima, T. & Kitamura, T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 (2000).
Neumann, G. et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl Acad. Sci. USA 96, 9345–9350 (1999).
Gorai, T. et al. F1Fo-ATPase, F-type proton-translocating ATPase, at the plasma membrane is critical for efficient influenza virus budding. Proc. Natl Acad. Sci. USA 109, 4615–4620 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682 (2012).
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Acknowledgements
The authors thank E. Takashita (National Institute of Infectious Diseases, Japan) for providing mouse anti-M1 (WS-27/52) and mouse anti-HA (WS3-54) antibodies, T. Kitamura (Institute of Medical Science, University of Tokyo) for providing Plat-GP cells and S. Watson for editing the manuscript. The authors thank T. Noda, E. Kawakami, T. Lopes, J. I-Hsuan Wang, Y. Sakai-Tagawa, K. Iwatsuki-Horimoto and our other co-workers for discussions and technical support. The authors also thank Y. Tomari (a grant-in-aid for Scientific Research on Innovative Areas, ‘Non-coding RNA neo-taxonomy’) for use of the super-resolution microscope and T. Watanabe (Carl Zeiss Microscopy) for help with microscope operation. This research was supported by the Japan Initiative for Global Research Network on Infectious Diseases from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and from the Japan Agency for Medical Research and Development (AMED); by grants-in-aid from the Ministry of Health, Labour and Welfare, Japan; by ERATO; by grants from the Strategic Basic Research Program of the Japan Science and Technology Agency; by the Advanced Research & Development Programs for Medical Innovation from AMED; and by JSPS, KAKENHI grant no. 15K19107. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
T.A., S.Y. and Y.K. conceived and designed the experiments. T.A. performed the experiments. T.A., S.Y., S.W., T.W. and Y.K. analysed the data. T.A. and Y.T. contributed materials and analysis tools. T.A., S.Y. and Y.K. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Figures 1-13. (PDF 2233 kb)
Rights and permissions
About this article
Cite this article
Ando, T., Yamayoshi, S., Tomita, Y. et al. The host protein CLUH participates in the subnuclear transport of influenza virus ribonucleoprotein complexes. Nat Microbiol 1, 16062 (2016). https://doi.org/10.1038/nmicrobiol.2016.62
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.62