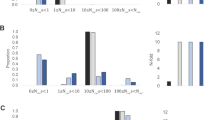
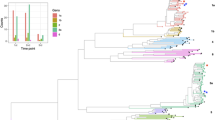
Abstract
Spontaneous mutations are the ultimate source of genetic variation and have a prominent role in evolution. RNA viruses such as hepatitis C virus (HCV) have extremely high mutation rates, but these rates have been inferred from a minute fraction of genome sites, limiting our view of how RNA viruses create diversity. Here, by applying high-fidelity ultradeep sequencing to a modified replicon system, we scored >15,000 spontaneous mutations, encompassing more than 90% of the HCV genome. This revealed >1,000-fold differences in mutability across genome sites, with extreme variations even between adjacent nucleotides. We identify base composition, the presence of high- and low-mutation clusters and transition/transversion biases as the main factors driving this heterogeneity. Furthermore, we find that mutability correlates with the ability of HCV to diversify in patients. These data provide a site-wise baseline for interrogating natural selection, genetic load and evolvability in HCV, as well as for evaluating drug resistance and immune evasion risks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Elde, N. C. et al. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 150, 831–841 (2012).
Goldberg, D. E., Siliciano, R. F. & Jacobs, W. R. Jr. Outwitting evolution: fighting drug-resistant TB, malaria, and HIV. Cell 148, 1271–1283 (2012).
Rosenberg, R. Detecting the emergence of novel, zoonotic viruses pathogenic to humans. Cell Mol. Life Sci. 72, 1115–1125 (2015).
Schotsaert, M. & Garcia-Sastre, A. Influenza vaccines: a moving interdisciplinary field. Viruses 6, 3809–3826 (2014).
Lauring, A. S., Frydman, J. & Andino, R. The role of mutational robustness in RNA virus evolution. Nature Rev. Microbiol. 11, 327–336 (2013).
Andino, R. & Domingo, E. Viral quasispecies. Virology 479–480, 46–51 (2015).
Menéndez-Arias, L. Mutation rates and intrinsic fidelity of retroviral reverse transcriptases. Viruses 1, 1137–1165 (2009).
Harris, R. S. & Dudley, J. P. APOBECs and virus restriction. Virology 479–480, 131–145 (2015).
Smith, E. C. & Denison, M. R. Coronaviruses as DNA wannabes: a new model for the regulation of RNA virus replication fidelity. PLoS Pathogens 9, e1003760 (2013).
Tomaselli, S., Galeano, F., Locatelli, F. & Gallo, A. ADARs and the balance game between virus infection and innate immune cell response. Curr. Issues Mol. Biol. 17, 37–52 (2015).
Sanjuán, R., Nebot, M. R., Chirico, N., Mansky, L. M. & Belshaw, R. Viral mutation rates. J. Virol. 84, 9733–9748 (2010).
Duffy, S., Shackelton, L. A. & Holmes, E. C. Rates of evolutionary change in viruses: patterns and determinants. Nature Rev. Genet. 9, 267–276 (2008).
Biek, R., Pybus, O. G., Lloyd-Smith, J. O. & Didelot, X. Measurably evolving pathogens in the genomic era. Trends Ecol. Evol. 30, 306–313 (2015).
Zhang, X., Rennick, L. J., Duprex, W. P. & Rima, B. K. Determination of spontaneous mutation frequencies in measles virus under nonselective conditions. J. Virol. 87, 2686–2692 (2013).
Gago, S., Elena, S. F., Flores, R. & Sanjuán, R. Extremely high mutation rate of a hammerhead viroid. Science 323, 1308 (2009).
Combe, M. & Sanjuán, R. Variation in RNA virus mutation rates across host cells. PLoS Pathogens 10, e1003855 (2014).
Cuevas, J. M., González-Candelas, F., Moya, A. & Sanjuán, R. The effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J. Virol. 83, 5760–5764 (2009).
Cuevas, J. M., Geller, R., Garijo, R., López-Aldeguer, J. & Sanjuán, R. Extremely high mutation rate of HIV-1 in vivo. PLoS Biol. 13, e1002251 (2015).
Ribeiro, R. M. et al. Quantifying the diversification of hepatitis C virus (HCV) during primary infection: estimates of the in vivo mutation rate. PLoS Pathogens 8, e1002881 (2012).
Acevedo, A., Brodsky, L. & Andino, R. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature 505, 686–690 (2014).
Gower, E., Estes, C., Blach, S., Razavi-Shearer, K. & Razavi, H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J. Hepatol. 61, S45–S57 (2014).
Westbrook, R. H. & Dusheiko, G. Natural history of hepatitis C. J. Hepatol. 61, S58–S68 (2014).
Farci, P. et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288, 339–344 (2000).
Pawlotsky, J. M. Treatment failure and resistance with direct-acting antiviral drugs against hepatitis C virus. Hepatology 53, 1742–1751 (2011).
Bartenschlager, R., Kaul, A. & Sparacio, S. Replication of the hepatitis C virus in cell culture. Antiviral Res. 60, 91–102 (2003).
Lohmann, V. et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285, 110–113 (1999).
Kennedy, S. R. et al. Detecting ultralow-frequency mutations by duplex sequencing. Nature Protoc. 9, 2586–2606 (2014).
Schmitt, M. W. et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. USA 109, 14508–14513 (2012).
Mauger, D. M. et al. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc. Natl Acad. Sci. USA 112, 3692–3697 (2015).
Bankwitz, D. et al. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J. Virol. 84, 5751–5763 (2010).
Taylor, D. R., Puig, M., Darnell, M. E., Mihalik, K. & Feinstone, S. M. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J. Virol. 79, 6291–6298 (2005).
Powdrill, M. H. et al. Contribution of a mutational bias in hepatitis C virus replication to the genetic barrier in the development of drug resistance. Proc. Natl Acad. Sci. USA 108, 20509–20513 (2011).
Bikard, D. & Marraffini, L. A. Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr. Opin. Immunol. 24, 15–20 (2012).
Guo, H., Arambula, D., Ghosh, P. & Miller, J. F. Diversity-generating retroelements in phage and bacterial genomes. Microbiol. Spectr. 2, MDNA3-0029-2014 (2014).
Geller, R. et al. The external domains of the HIV-1 envelope are a mutational cold spot. Nature Commun. 6, 8571 (2015).
Duchène, S., Ho, S. Y. & Holmes, E. C. Declining transition/transversion ratios through time reveal limitations to the accuracy of nucleotide substitution models. BMC Evol. Biol. 15, 36 (2015).
Hershberg, R. & Petrov, D. A. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6, e1001115 (2010).
Zhang, Z. & Gerstein, M. Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res. 31, 5338–5348 (2003).
Keller, I., Bensasson, D. & Nichols, R. A. Transition–transversion bias is not universal: a counter example from grasshopper pseudogenes. PLoS Genet. 3, e22 (2007).
Zhu, Y. O., Siegal, M. L., Hall, D. W. & Petrov, D. A. Precise estimates of mutation rate and spectrum in yeast. Proc. Natl Acad. Sci. USA 111, E2310–E2318 (2014).
Ossowski, S. et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327, 92–94 (2010).
Pause, A. et al. An NS3 serine protease inhibitor abrogates replication of subgenomic hepatitis C virus RNA. J. Biol. Chem. 278, 20374–20380 (2003).
Pietschmann, T., Lohmann, V., Rutter, G., Kurpanek, K. & Bartenschlager, R. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75, 1252–1264 (2001).
Binder, M. et al. Replication vesicles are load- and choke-points in the hepatitis C virus lifecycle. PLoS Pathogens 9, e1003561 (2013).
Gosert, R. et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77, 5487–5492 (2003).
Quinkert, D., Bartenschlager, R. & Lohmann, V. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79, 13594–13605 (2005).
Moradpour, D., Penin, F. & Rice, C. M. Replication of hepatitis C virus. Nature Rev. Microbiol. 5, 453–463 (2007).
Dahari, H., Ribeiro, R. M., Rice, C. M. & Perelson, A. S. Mathematical modeling of subgenomic hepatitis C virus replication in Huh-7 cells. J. Virol. 81, 750–760 (2007).
Acknowledgements
The authors thank R. Bartenschlager for the HCV replicon, F. González-Candelas for the viral RNA, V. Sentandreu and A. Martínez for help with optimization of the Duplex Sequencing technique, R. Flores for suggestions, M. Binder for discussions on HCV replication models and S. Torres for laboratory assistance. This work was financially supported by grants from the European Research Council (ERC-2011-StG- 281191-VIRMUT) and the Spanish MINECO (BFU2013-41329) to R.San.
Author information
Authors and Affiliations
Contributions
R.Ge. performed the experiments, including replicon construction, passaging, Sanger and Duplex Sequencing, and performed the bioinformatics analysis. U.E. and J.M.C. contributed to optimizing the Duplex Sequencing. I.A., J.-V.B. and J.M.C. performed Sanger sequencing of the replicon. R.Ga. performed Sanger sequencing of in vitro polymerization products. R.Sab., J.B.P. and A.M. performed in vitro polymerization assays. R.San. designed and supervised research, acquired funding, analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Tables 1-2, Figures 1-4 and References (PDF 5452 kb)
Supplementary Dataset 1
Sequence of the seven cloned fragments and the fragment used for in vitro polymerization. (TXT 10 kb)
Supplementary Dataset 2
List of mutations obtained by Duplex Sequencing in the three replicon lines, mapped to annotated H77 reference sequence including sequencing coverage, patient sequence diversity, and SHAPE data. (XLSX 2594 kb)
Rights and permissions
About this article
Cite this article
Geller, R., Estada, Ú., Peris, J. et al. Highly heterogeneous mutation rates in the hepatitis C virus genome. Nat Microbiol 1, 16045 (2016). https://doi.org/10.1038/nmicrobiol.2016.45
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.45
This article is cited by
-
Mutational spectrum of hepatitis C virus in patients with chronic hepatitis C determined by single molecule real-time sequencing
Scientific Reports (2022)
-
Increased RNA virus population diversity improves adaptability
Scientific Reports (2021)
-
Analysis of drug-resistance-associated mutations and genetic barriers in hepatitis C virus NS5B sequences in China
Archives of Virology (2020)
-
Hepatitis C virus sequence divergence preserves p7 viroporin structural and dynamic features
Scientific Reports (2019)
-
HA1077 displays synergistic activity with daclatasvir against hepatitis C virus and suppresses the emergence of NS5A resistance-associated substitutions in mice
Scientific Reports (2018)