Abstract
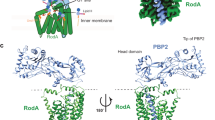
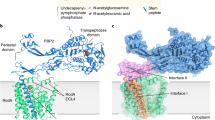
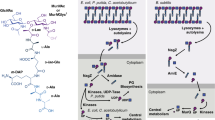
The bacterial cell wall is a highly conserved essential component of most bacterial groups. It is the target for our most frequently used antibiotics and provides important small molecules that trigger powerful innate immune responses. The wall is composed of glycan strands crosslinked by short peptides. For many years, the penicillin-binding proteins were thought to be the key enzymes required for wall synthesis. RodA and possibly other proteins in the wider SEDS (shape, elongation, division and sporulation) family have now emerged as a previously unknown class of essential glycosyltranferase enzymes, which play key morphogenetic roles in bacterial cell wall synthesis. We provide evidence in support of this role and the discovery of small natural product molecules that probably target these enzymes. The SEDS proteins have exceptional potential as targets for new antibacterial therapeutic agents.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Errington, J. L-form bacteria, cell walls and the origins of life. Open Biol. 3, 120143 (2013).
Typas, A., Banzhaf, M., Gross, C. A. & Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 (2012).
Lovering, A. L., Safadi, S. S. & Strynadka, N. C. J. Structural perspective of peptidoglycan biosynthesis and assembly. Annu. Rev. Biochem. 81, 451–478 (2012).
Spratt, B. G. Distinct penicillin-binding proteins involved in the division, elongation and shape of Escherichia coli K-12. Proc. Natl Acad. Sci. USA 72, 2999–3003 (1975).
Wei, Y., Havasy, T., McPherson, D. C. & Popham, D. L. Rod shape determination by the Bacillus subtilis class B penicillin-binding proteins encoded by pbpA and pbpH. J. Bacteriol. 185, 4717–4726 (2003).
Daniel, R. A., Williams, A. M. & Errington, J. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J. Bacteriol. 178, 2343–2350 (1996).
McPherson, D. C. & Popham, D. L. Peptidoglycan synthesis in the absence of class a penicillin-binding proteins in Bacillus subtilis. J. Bacteriol. 185, 1423–1431 (2003).
Henriques, A. O., Glaser, P., Piggot, P. J. & Moran, C. P. Jr. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28, 235–247 (1998).
Meeske, A. J. et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016).
Cho, H. et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol. 1, 16172 (2016).
van Heijenoort, Y., Leduc, M., Singer, H. & van Heijenoort, J. Effects of moenomycin on Escherichia coli. J. Gen. Microbiol. 133, 667–674 (1987).
Eiamphungporn, W. & Helmann, J. D. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67, 830–848 (2008).
Salzberg, L. I., Luo, Y., Hachmann, A. B., Mascher, T. & Helmann, J. D. The Bacillus subtilis GntR family repressor YtrA responds to cell wall antibiotics. J. Bacteriol. 193, 5793–5801 (2011).
McPherson, D. C., Driks, A. & Popham, D. L. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J. Bacteriol. 183, 6046–6053 (2001).
Vasudevan, P., McElligott, J., Attkisson, C., Betteken, M. & Popham, D. L. Homologues of the Bacillus subtilis SpoVB protein are involved in cell wall metabolism. J. Bacteriol. 191, 6012–6019 (2009).
Meeske, A. J. et al. Murj and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl Acad. Sci. USA 112, 6437–6442 (2015).
Adams, D. W. & Errington, J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642–653 (2009).
Wachi, M., Doi, M., Okada, Y. & Matsuhashi, M. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J. Bacteriol. 171, 6511–6516 (1989).
Leaver, M. & Errington, J. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol. Microbiol. 57, 1196–1209 (2005).
Kawai, Y. et al. A widespread family of bacterial cell wall assembly proteins. EMBO J. 30, 4931–4941 (2011).
Kawai, Y., Daniel, R. A. & Errington, J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol. Microbiol. 71, 1131–1144 (2009).
Errington, J. Bacterial morphogenesis and the enigmatic MreB helix. Nat. Rev. Microbiol. 13, 241–248 (2015).
Ikeda, M. et al. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 171, 6375–6378 (1989).
Fay, A., Meyer, P. & Dworkin, J. Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J. Mol. Biol. 399, 547–561 (2010).
Tamaki, S., Matsuzawa, H. & Matsuhashi, M. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J. Bacteriol. 141, 52–57 (1980).
Höltje, J. V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62, 181–203 (1998).
Sham, L. T. et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–222 (2014).
Ovchinnikov, S. et al. Large-scale determination of previously unsolved protein structures using evolutionary information. eLife 4, e09248 (2015).
Maguin, E., Prévost, H., Ehrlich, S. D. & Gruss, A. Efficient insertional mutagenesis in lactococci and other Gram-positive bacteria. J. Bacteriol. 178, 931–935 (1996).
Claessen, D. et al. Control of the cell elongation–division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 68, 1029–1046 (2008).
Glaser, P. et al. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11, 1160–1168 (1997).
Acknowledgements
This work was funded by grants from the BBSRC (BB/G015902/1, to R.A.D., J.E. and A.G.), ERC (670980, to J.E., Y.K. and K.E.) and the Wellcome Trust (WT098374AIA, to J.E. and L.J.W.) and core resources in Demuris Ltd. The authors thank R. Emmins, who constructed some of the mutants, D. Roberts for help with microfluidic microscopy and R. Sweet for help with the large-scale fermentation run.
Author information
Authors and Affiliations
Contributions
K.E., A.G., Y.K., J.D., L.J.W. and R.A.D. carried out the experiments. All authors contributed to the experimental design and concepts, and that all authors contributed to the text. J.E. wrote the main text with contributions from all other authors.
Corresponding authors
Ethics declarations
Competing interests
J.E. is a director and shareholder at Demuris Ltd. N.A. and J.D. are employees at Demuris Ltd.
Supplementary information
Supplementary Information
Legends for all Supplementary Material and Supplementary Figures 2–5. (PDF 795 kb)
Supplementary Figures and Tables
Supplementary Figure 1 and Supplementary Tables 1 and 2. (PDF 845 kb)
Supplementary Video 1
Effect of compound 654/A (30 μl) on growth and morphology of the Δ4 strain. Approximately 3 hours after addition of 654/A to Δ4 cells in the microfluidic device, cell growth slows down significantly and extensive cell lysis occurs. (AVI 19450 kb)
Supplementary Video 2
Untreated control for growth and morphology of the Δ4 strain. In the absence of compound 654/A, Δ4 cells continue their normal growth up to full confluency. (AVI 39740 kb)
Supplementary Video 3
Effect of compound 654/A (30 μl) on growth and morphology of the wild type. Wild-type cells show normal growth at relatively low concentrations of 654/A. (AVI 34112 kb)
Supplementary Video 4
Effect of compound 654/A (70 μl) on growth and morphology of the wild type. At relatively high concentrations of 654/A, extensive lysis of wild-type cells occurs, similar to that of the Δ4 cells (see Supplementary Video 1). (AVI 25887 kb)
Rights and permissions
About this article
Cite this article
Emami, K., Guyet, A., Kawai, Y. et al. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol 2, 16253 (2017). https://doi.org/10.1038/nmicrobiol.2016.253
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.253
This article is cited by
-
Cell constriction requires processive septal peptidoglycan synthase movement independent of FtsZ treadmilling in Staphylococcus aureus
Nature Microbiology (2024)
-
Diversity of sugar-diphospholipid-utilizing glycosyltransferase families
Communications Biology (2024)
-
Coordinated peptidoglycan synthases and hydrolases stabilize the bacterial cell wall
Nature Communications (2023)
-
Evidence of two differentially regulated elongasomes in Salmonella
Communications Biology (2023)
-
On the mechanisms of lysis triggered by perturbations of bacterial cell wall biosynthesis
Nature Communications (2023)