Abstract
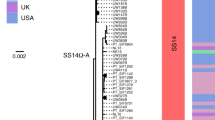
The abrupt onslaught of the syphilis pandemic that started in the late fifteenth century established this devastating infectious disease as one of the most feared in human history1. Surprisingly, despite the availability of effective antibiotic treatment since the mid-twentieth century, this bacterial infection, which is caused by Treponema pallidum subsp. pallidum (TPA), has been re-emerging globally in the last few decades with an estimated 10.6 million cases in 2008 (ref. 2). Although resistance to penicillin has not yet been identified, an increasing number of strains fail to respond to the second-line antibiotic azithromycin3. Little is known about the genetic patterns in current infections or the evolutionary origins of the disease due to the low quantities of treponemal DNA in clinical samples and difficulties in cultivating the pathogen4. Here, we used DNA capture and whole-genome sequencing to successfully interrogate genome-wide variation from syphilis patient specimens, combined with laboratory samples of TPA and two other subspecies. Phylogenetic comparisons based on the sequenced genomes indicate that the TPA strains examined share a common ancestor after the fifteenth century, within the early modern era. Moreover, most contemporary strains are azithromycin-resistant and are members of a globally dominant cluster, named here as SS14-Ω. The cluster diversified from a common ancestor in the mid-twentieth century subsequent to the discovery of antibiotics. Its recent phylogenetic divergence and global presence point to the emergence of a pandemic strain cluster.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gall, G. E. C., Lautenschlager, S. & Bagheri, H. C. Quarantine as a public health measure against an emerging infectious disease: syphilis in Zurich at the dawn of the modern era (1496–1585). GMS Hyg. Infect. Control 11, 13 (2016).
Rowley, J. et al. Global Incidence and Prevalence of Selected Curable Sexually Transmitted Infections, 2008 (World Health Organization, 2012).
Stamm, L. V. Global challenge of antibiotic-resistant Treponema pallidum. Antimicrob. Agents Chemother. 54, 583–589 (2010).
Fraser, C. M. et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281, 375–388 (1998).
Quétel, C. History of Syphilis (Johns Hopkins Univ. Press, 1990).
Fernandez de Oviedo y Valdes, G. Sumario de la natural historia de las Indias (Fondo de Cultura Economico, 1526).
Harper, K. N., Zuckerman, M. K., Harper, M. L., Kingston, J. D. & Armelagos, G. J. The origin and antiquity of syphilis revisited: an appraisal of Old World pre-Columbian evidence for treponemal infection. Am. J. Phys. Anthropol. 146(Suppl 53), 99–133 (2011).
Šmajs, D., Norris, S. J. & Weinstock, G. M. Genetic diversity in Treponema pallidum: implications for pathogenesis, evolution and molecular diagnostics of syphilis and yaws. Infect. Genet. Evol. 12, 191–202 (2012).
Giacani, L. et al. Complete genome sequence of the Treponema pallidum subsp. pallidum Sea81-4 strain. Genome Announc. 2, e00333-14 (2014).
Štaudová, B. et al. Whole genome sequence of the Treponema pallidum subsp. endemicum strain Bosnia A: the genome is related to yaws treponemes but contains few loci similar to syphilis treponemes. PLoS Negl. Trop. Dis. 8, e3261 (2014).
Marra, C. M. et al. Enhanced molecular typing of Treponema pallidum: geographical distribution of strain types and association with neurosyphilis. J. Infect. Dis. 202, 1380–1388 (2010).
Grillová, L. et al. Molecular typing of Treponema pallidum in the Czech Republic during 2011 to 2013: increased prevalence of identified genotypes and of isolates with macrolide resistance. J. Clin. Microbiol. 52, 3693–3700 (2014).
Hodges, E. et al. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat. Protoc. 4, 960–974 (2009).
Meyer, M. & Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. http://dx.doi.org/10.1101/pdb.prot5448 (2010).
Pětrošová, H. et al. Resequencing of Treponema pallidum ssp. pallidum strains Nichols and SS14: correction of sequencing errors resulted in increased separation of syphilis treponeme subclusters. PLoS ONE 8, e74319 (2013).
Centurion-Lara, A. et al. Fine analysis of genetic diversity of the tpr gene family among treponemal species, subspecies and strains. PLoS Negl. Trop. Dis. 7, e2222 (2013).
Mikalova, L. et al. Genome analysis of Treponema pallidum subsp. pallidum and subsp. pertenue strains: most of the genetic differences are localized in six regions. PLoS ONE 5, e15713 (2010).
Achtman, M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 62, 53–70 (2008).
Pětrošová, H. et al. Whole genome sequence of Treponema pallidum ssp. pallidum, strain Mexico A, suggests recombination between yaws and syphilis strains. PLoS Negl. Trop. Dis. 6, e1832 (2012).
Bouckaert, R. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014).
Lukehart, S. A. & Giacani, L. When is syphilis not syphilis? Or is it? Sex. Transm. Dis. 41, 554–555 (2014).
Stadler, T., Kuhnert, D., Bonhoeffer, S. & Drummond, A. J. Birth–death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV). Proc. Natl Acad. Sci. USA 110, 228–233 (2013).
Holt, K. E. et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat. Genet. 44, 1056–1059 (2012).
Mutreja, A. et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477, 462–465 (2011).
Giacani, L. et al. Footprint of positive selection in Treponema pallidum subsp. pallidum genome sequences suggests adaptive microevolution of the syphilis pathogen. PLoS Negl. Trop. Dis. 6, e1698 (2012).
Strouhal, M. et al. Genome differences between Treponema pallidum subsp. pallidum strain Nichols and T. paraluiscuniculi strain Cuniculi A. Infect. Immun. 75, 5859–5866 (2007).
Marra, C. M. et al. Antibiotic selection may contribute to increases in macrolide-resistant Treponema pallidum. J. Infect. Dis. 194, 1771–1773 (2006).
Mitchell, S. J. et al. Azithromycin-resistant syphilis infection: San Francisco, California, 2000–2004. Clin. Infect. Dis. 42, 337–345 (2006).
Morshed, M. & Jones, H. Treponema pallidum macrolide resistance in BC. Can. Med. Assoc. J. 174, 349 (2006).
Matejkova, P. et al. Macrolide treatment failure in a case of secondary syphilis: a novel A2059G mutation in the 23S rRNA gene of Treponema pallidum subsp. pallidum. J. Med. Microbiol. 58, 832–836 (2009).
Stamm, L. V. & Bergen, H. L. A point mutation associated with bacterial macrolide resistance is present in both 23S rRNA genes of an erythromycin-resistant Treponema pallidum clinical isolate. Antimicrob. Agents Chemother. 44, 806–807 (2000).
Šmajs, D., Paštěková, L. & Grillová, L. Macrolide resistance in the syphilis spirochete, Treponema pallidum ssp. pallidum: can we also expect macrolide-resistant yaws strains? Am. J. Trop. Med. Hyg. 93, 678–683 (2015).
Geisler, W. M. et al. Azithromycin versus doxycycline for urogenital Chlamydia trachomatis infection. N. Engl. J. Med. 373, 2512–2521 (2015).
Centurion-Lara, A. in Pathogenic Treponema: Molecular and Cellular Biology (eds Radolf, J. D. & Lukehart, S. A. ) 267–283 (Caister Academic, 2006).
Knell, R. J. Syphilis in renaissance Europe: rapid evolution of an introduced sexually transmitted disease? Proc. Biol. Sci. 271(Suppl 4), S174–S176 (2004).
Kircher, M., Sawyer, S. & Meyer, M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2012).
Peltzer, A. et al. EAGER: Efficient ancient genome reconstruction. Genome Biol. 17, 60 (2016).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Simpson, J. T. & Durbin, R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 22, 549–556 (2012).
Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002).
Strimmer, K. & von Haeseler, A. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13, 964–969 (1996).
Strimmer, K. & Rambaut, A. Inferring confidence sets of possibly misspecified gene trees. Proc. R. Soc. B 269, 137–142 (2002).
Shimodaira, H. & Hasegawa, M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16, 1114–1116 (1999).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Librado, P. & Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Baele, G. et al. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 29, 2157–2167 (2012).
Rambaut, A., Suchard, M., Xie, D. & Drummond, A. Tracer v1.6. (2014); http://beast.bio.ed.ac.uk/Tracer
Rambaut, A. FigTree v.1.4.2. (2014); http://tree.bio.ed.ac.uk/software/figtree/
Bandelt, H.-J., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Dai, T. et al. Molecular typing of Treponema pallidum: a 5-year surveillance in Shanghai, China. J. Clin. Microbiol. 50, 3674–3677 (2012).
Flasarová, M. et al. Sequencing-based molecular typing of Treponema pallidum strains in the Czech Republic: all identified genotypes are related to the sequence of the SS14 strain. Acta Derm. Venereol. 92, 669–674 (2012).
Grange, P. A. et al. Molecular subtyping of Treponema pallidum in Paris, France. Sex. Transm. Dis. 40, 641–644 (2013).
Grimes, M. et al. Two mutations associated with macrolide resistance in Treponema pallidum: increasing prevalence and correlation with molecular strain type in Seattle, Washington. Sex. Transm. Dis. 39, 954–958 (2012).
Peng, R.-R. et al. Molecular typing of Treponema pallidum causing early syphilis in China: a cross-sectional study. Sex. Transm. Dis. 39, 42–45 (2012).
Tian, H. et al. Molecular typing of Treponema pallidum: identification of a new sequence of tp0548 gene in Shandong, China. Sex. Transm. Dis. 41, 551 (2014).
Tipple, C., McClure, M. O. & Taylor, G. P. High prevalence of macrolide resistant Treponema pallidum strains in a London centre. Sex. Transm. Infect. 87, 486–488 (2011).
Wu, B.-R. et al. Multicentre surveillance of prevalence of the 23S rRNA A2058G and A2059G point mutations and molecular subtypes of Treponema pallidum in Taiwan, 2009–2013. Clin. Microbiol. Infect. 20, 802–807 (2014).
Acknowledgements
Research in Zurich by N.A. and H.C.B. was funded by the Forschungskredit and the University of Zurich. A.H. was funded by an ERC Starting Grant. F.G.C. and L.S.B. were funded by MINECO (Spanish Government) and PROMETEO (Generalitat Valenciana). K.I.B. was funded by the Social Sciences and Humanities Research Council of Canada. L.M. was funded by the Faculty of Medicine of Masaryk University. The authors thank S. Lautenschlager for guidance, A. Drummond for input on BEAST, S. Lukehart for providing HaitiB, Sea86-1, Bal3, Bal9, Bal73-1 and Grady1 strain DNA, and C. Marra for providing UW249B and UW231B strain DNA. The authors also thank A. Messina and the S3IT at the University of Zurich for providing computational resources and services, and I. Schoechli and L. Keller's group for their valued support.
Author information
Authors and Affiliations
Contributions
N.A. and H.C.B. conceived the investigation. N.A., L.G., S.J.N., D.S., P.P.B., F.G.-C., K.N., J.K. and H.C.B. devised research and analyses. N.A., G.J., A.P., A.S., A.H., M.S., L.G., L.S.-B., D.K., L.R.D., L.M., F.G.-C. and K.N. analysed data. N.A., V.J.S., M.S., L.G., K.I.B., L.R.D., L.G.V. and P.P.B. contributed to or performed experiments. M.S., L.G., S.B., P.K., P.F., P.R.G., M.A.P., L.G.V., M.R.F., A.M., D.S., P.P.B. and F.G.-C. provided clinical samples and A.C.L., L.G., S.J.N. and D.S. provided laboratory samples. N.A. and H.C.B. wrote the manuscript with significant contributions from M.S., L.G., L.S.-B., D.K., K.I.B., L.R.D., L.M., S.B., L.G., S.J.N., D.S., P.P.B., F.G.-C., K.N. and J.K. and with comments from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Methods; Supplementary Tables 3, 5, 6, 9, 10; Legends for all Supplementary Tables; Supplementary Figures 1-6; Supplementary References. (PDF 820 kb)
Supplementary Table 1
Sample Information. (XLSX 14 kb)
Supplementary Table 2
Read preprocessing, mapping and genotyping results. (XLSX 23 kb)
Supplementary Table 4
SNP calls for samples used in genome-wide data analyses (n = 39). (XLSX 14 kb)
Supplementary Table 7
Putative recombinant genes identified by Gubbins and ClonalFrameML. (XLSX 16 kb)
Supplementary Table 8
Clade classification and mutations associated with antibiotic resistance (for all sequenced and published samples). (XLSX 12 kb)
Rights and permissions
About this article
Cite this article
Arora, N., Schuenemann, V., Jäger, G. et al. Origin of modern syphilis and emergence of a pandemic Treponema pallidum cluster. Nat Microbiol 2, 16245 (2017). https://doi.org/10.1038/nmicrobiol.2016.245
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.245
This article is cited by
-
Redefining the treponemal history through pre-Columbian genomes from Brazil
Nature (2024)
-
Resurgence of syphilis: focusing on emerging clinical strategies and preclinical models
Journal of Translational Medicine (2023)
-
Core genome sequencing and genotyping of Leptospira interrogans in clinical samples by target capture sequencing
BMC Infectious Diseases (2023)
-
A suite of PCR-LwCas13a assays for detection and genotyping of Treponema pallidum in clinical samples
Nature Communications (2022)
-
High frequency of Nichols-like strains and increased levels of macrolide resistance in Treponema pallidum in clinical samples from Buenos Aires, Argentina
Scientific Reports (2022)