Abstract
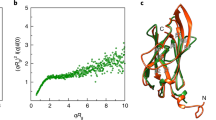
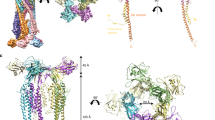
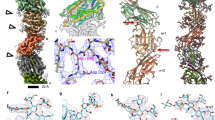
Archaea use flagella known as archaella—distinct both in protein composition and structure from bacterial flagella—to drive cell motility, but the structural basis of this function is unknown. Here, we report an atomic model of the archaella, based on the cryo electron microscopy (cryoEM) structure of the Methanospirillum hungatei archaellum at 3.4 Å resolution. Each archaellum contains ∼61,500 archaellin subunits organized into a curved helix with a diameter of 10 nm and average length of 10,000 nm. The tadpole-shaped archaellin monomer has two domains, a β-barrel domain and a long, mildly kinked α-helix tail. Our structure reveals multiple post-translational modifications to the archaella, including six O-linked glycans and an unusual N-linked modification. The extensive interactions among neighbouring archaellins explain how the long but thin archaellum maintains the structural integrity required for motility-driving rotation. These extensive inter-subunit interactions and the absence of a central pore in the archaellum distinguish it from both the bacterial flagellum and type IV pili.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
14 July 2017
In the PDF version of this article previously published, the year of publication provided in the footer of each page and in the 'How to cite' section was erroneously given as 2017, it should have been 2016. This error has now been corrected. The HTML version of the article was not affected.
References
Albers, S.-V. & Jarrell, K. F. The archaellum: how archaea swim. Front. Microbiol. 6, 23 (2015).
Näther, D. J., Rachel, R., Wanner, G. & Wirth, R. Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell–cell contacts. J. Bacteriol. 188, 6915–6923 (2006).
Szabó, Z. et al. Flagellar motility and structure in the hyperthermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 189, 4305–4309 (2007).
Jarrell, K. F. & Albers, S.-V. The archaellum: an old motility structure with a new name. Trends Microbiol. 20, 307–312 (2012).
Bellack, A., Huber, H., Rachel, R., Wanner, G. & Wirth, R. Methanocaldococcus villosus sp. nov., a heavily flagellated archaeon that adheres to surfaces and forms cell–cell contacts. Int. J. Syst. Evol. Micr. 61, 1239–1245 (2011).
Jarrell, K. F., Stark, M., Nair, D. B. & Chong, J. P. J. Flagella and pili are both necessary for efficient attachment of Methanococcus maripaludis to surfaces. FEMS Microbiol. Lett. 319, 44–50 (2011).
Schopf, S., Wanner, G., Rachel, R. & Wirth, R. An archaeal bi-species biofilm formed by Pyrococcus furiosus and Methanopyrus kandleri. Arch. Microbiol. 190, 371–377 (2008).
Alam, M. & Oesterhelt, D. Morphology, function and isolation of halobacterial flagella. J. Mol. Biol. 176, 459–475 (1984).
Faguy, D. M., Jarrell, K. F., Kuzio, J. & Kalmokoff, M. L. Molecular analysis of archaeal flagellins: similarity to the type IV pilin-transport superfamily widespread in bacteria. Can. J. Microbiol. 40, 67–71 (1994).
Correia, J. D. & Jarrell, K. F. Posttranslational processing of Methanococcus voltae preflagellin by preflagellin peptidases of M. voltae and other methanogens. J. Bacteriol. 182, 855–858 (2000).
Patenge, N., Berendes, A., Engelhardt, H., Schuster, S. C. & Oesterhelt, D. The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum. Mol. Microbiol. 41, 653–663 (2001).
Thomas, N. A., Pawson, C. T. & Jarrell, K. F. Insertional inactivation of the flaH gene in the archaeon Methanococcus voltae results in non-flagellated cells. Mol. Genet. Genomics 265, 596–603 (2001).
Chaban, B. et al. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol. Microbiol. 66, 596–609 (2007).
Lassak, K. et al. Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83, 110–124 (2012).
Streif, S., Staudinger, W. F., Marwan, W. & Oesterhelt, D. Flagellar rotation in the archaeon Halobacterium salinarum depends on ATP. J. Mol. Biol. 384, 1–8 (2008).
Banerjee, A. et al. Flaf is a β-sandwich protein that anchors the archaellum in the archaeal cell envelope by binding the S-layer protein. Structure 23, 863–872 (2015).
Chaudhury, P. et al. The nucleotide-dependent interaction of FlaH and FlaI is essential for assembly and function of the archaellum motor. Mol. Microbiol. 99, 674–685 (2016).
Trachtenberg, S., Galkin, V. E. & Egelman, E. H. Refining the structure of the Halobacterium salinarum flagellar filament using the iterative helical real space reconstruction method: insights into polymorphism. J. Mol. Biol. 346, 665–676 (2005).
Trachtenberg, S. & Cohen-Krausz, S. The archaeabacterial flagellar filament: a bacterial propeller with a pilus-like structure. J. Mol. Microb Biotech. 11, 208–220 (2006).
Beveridge, T. J., Sprott, G. D. & Whippey, P. Ultrastructure, inferred porosity, and Gram-staining character of Methanospirillum hungatei filament termini describe a unique cell permeability for this archaeobacterium. J. Bacteriol. 173, 130–140 (1991).
Xu, W. et al. Modeling and measuring the elastic properties of an archaeal surface, the sheath of Methanospirillum hungatei, and the implication of methane production. J. Bacteriol. 178, 3106–3112 (1996).
Toso, D. B., Henstra, A. M., Gunsalus, R. P. & Zhou, Z. H. Structural, mass and elemental analyses of storage granules in methanogenic archaeal cells. Environ. Microbiol. 13, 2587–2599 (2011).
Gunsalus, R. P. et al. Complete genome sequence of Methanospirillum hungatei type strain JF1. Standards Genomic Sci. 11, 2 (2016).
Bardy, S. L. & Jarrell, K. F. Flak of the archaeon Methanococcus maripaludis possesses preflagellin peptidase activity. FEMS Microbiol. Lett. 208, 53–59 (2002).
Bardy, S. L. & Jarrell, K. F. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol. Microbiol. 50, 1339–1347 (2003).
Szabó, Z. et al. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 189, 772–778 (2007).
Albers, S.-V., Szabó, Z. & Driessen, A. J. M. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 185, 3918–3925 (2003).
Ng, S. Y. M., Chaban, B. & Jarrell, K. F. Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J. Mol. Microbiol. Biotech. 11, 167–191 (2006).
Kelly, J., Logan, S. M., Jarrell, K. F., VanDyke, D. J. & Vinogradov, E. A novel N-linked flagellar glycan from Methanococcus maripaludis. Carbohyd. Res. 344, 648–653 (2009).
Voisin, S. et al. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J. Biol. Chem. 280, 16586–16593 (2005).
Sun, S. & Zhang, H. Identification and validation of atypical N-glycosylation sites. Anal. Chem. 87, 11948–11951 (2015).
Banerjee, A., Neiner, T., Tripp, P. & Albers, S.-V. Insights into subunit interactions in the Sulfolobus acidocaldarius archaellum cytoplasmic complex. FEBS J. 280, 6141–6149 (2013).
Jarrell, K. F., Bayley, D. P. & Kostyukova, A. S. The archaeal flagellum: a unique motility structure. J. Bacteriol. 178, 5057–5064 (1996).
Tripepi, M. et al. N-glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. J. Bacteriol. 194, 4876–4887 (2012).
Ding, Y. et al. Effects of N-glycosylation site removal in archaellins on the assembly and function of archaella in Methanococcus maripaludis. PLoS ONE 10, e0116402 (2015).
Meyer, B. H. et al. Agl16, a thermophilic glycosyltransferase mediating the last step of N-glycan biosynthesis in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J. Bacteriol. 195, 2177–2186 (2013).
Meyer, B. H., Birich, A. & Albers, S.-V. N-glycosylation of the archaellum filament is not important for archaella assembly and motility, although N-glycosylation is essential for motility in Sulfolobus acidocaldarius. Biochimie 118, 294–301 (2015).
Turner, L., Ryu, W. S. & Berg, H. C. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182, 2793–2801 (2000).
Yonekura, K., Maki-Yonekura, S. & Namba, K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424, 643–650 (2003).
Craig, L. et al. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23, 651–662 (2006).
Mortezaei, N. et al. Structure and function of enterotoxigenic Escherichia coli fimbriae from differing assembly pathways. Mol. Microbiol. 95, 116–126 (2015).
Silverman, P. M. Towards a structural biology of bacterial conjugation. Mol. Microbiol. 23, 423–429 (1997).
McLaughlin, L. S., Haft, R. J. F. & Forest, K. T. Structural insights into the type II secretion nanomachine. Curr. Opin. Struc. Biol. 22, 208–216 (2012).
Yu, X. et al. Filaments from Ignicoccus hospitalis show diversity of packing in proteins containing N-terminal type IV pilin helices. J. Mol. Biol. 422, 274–281 (2012).
Henche, A.-L. et al. Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius. Environ. Microbiol. 14, 3188–3202 (2012).
Braun, T. et al. Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain. Proc. Natl Acad. Sci. USA 113, 10352–10357 (2016).
Faguy, D. M., Koval, S. F. & Jarrell, K. F. Effect of changes in mineral composition and growth temperature on filament length and flagellation in the Archaeon Methanospirillum hungatei. Arch. Microbiol. 159, 512–520 (1993).
Patel, G. B., Roth, L. A., Berg, L. v. d. & Clark, D. S. Characterization of a strain of Methanospirillum hungatii. Can. J. Microbiol. 22, 1404–1410 (1976).
Edman, P. & Begg, G. A protein sequenator. Eur. J. Biochem. 1, 80–91 (1967).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Altschul, S. F. et al. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109 (2005).
Jones, D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 (1999).
Suloway, C. et al. Fully automated, sequential tilt-series acquisition with Leginon. J. Struct. Biol. 167, 11–18 (2009).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013).
DeRosier, D. J. & Moore, P. B. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J. Mol. Biol. 52, 355–369 (1970).
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Egelman, E. H. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J. Struct. Biol. 157, 83–94 (2007).
Ge, P. et al. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 327, 689–693 (2010).
Scheres, S. H. W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Clemens, D. L., Ge, P., Lee, B.-Y., Horwitz, M. A. & Zhou, Z. H. Atomic structure of T6SS reveals interlaced array essential to function. Cell 160, 940–951 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Pettersen, E. F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
This project received support from National Institutes of Health grants GM071940 and AI094386, NIH/NCRR/NCATS UCLA CTSI grant UL1TR000124, from the UCLA-DOE Institute (DE-FC03-02ER6342) to R.P.G. and R.O.L., and NSF grants DMR-1548924 to Z.H.Z. and 1515843 to R.P.G. N.P. was supported in part by the NIH Biotechnology Training Grant Program (T32GM067555). P.G. was supported in part by an American Heart Association Western States Affiliates Postdoc Fellowship (13POST17340020). The authors acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from NIH (1S10OD018111) and NSF (DBI-1338135). NIH support for mass spectrometry was provided by grant S10RR025600. The authors acknowledge computer time at the Extreme Science and Engineering Discovery Environment (XSEDE, grant MCB140140 to Z.H.Z.).
Author information
Authors and Affiliations
Contributions
Z.H.Z. and R.P.G. designed the project. N.P., P.G., R.R.O.L. and H.H.N. performed the experiments and analysed the data. Z.H.Z., R.P.G. and N.P. wrote the paper. All authors contributed to editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Video Legends 1–8, Supplementary Figures 1-9, Supplementary Tables 1–3. (PDF 2758 kb)
Supplementary Video 1
Overview of the M. hungatei cryoEM density map. (AVI 21147 kb)
Supplementary Video 2
Overview of the M. hungatei FlaB monomer model. (AVI 32286 kb)
Supplementary Video 3
Overview of the extra densities in the cryoEM map. (AVI 26432 kb)
Supplementary Video 4
Overview of inter-subunit interactions. (AVI 37145 kb)
Supplementary Video 5
Focus on inter-subunit hydrophobic interactions. (AVI 29729 kb)
Supplementary Video 6
Focus on inter-subunit ionic interactions. (AVI 11128 kb)
Supplementary Video 7
Comparison between a bacterial pilin, archaellin and bacterial flagellin. (AVI 17338 kb)
Supplementary Video 8
Comparison between protofilaments of bacterial pili, archaella and bacterial flagella. (AVI 11914 kb)
Rights and permissions
About this article
Cite this article
Poweleit, N., Ge, P., Nguyen, H. et al. CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pilus. Nat Microbiol 2, 16222 (2017). https://doi.org/10.1038/nmicrobiol.2016.222
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.222
This article is cited by
-
Methanogenic partner influences cell aggregation and signalling of Syntrophobacterium fumaroxidans
Applied Microbiology and Biotechnology (2024)
-
Hierarchical organization and assembly of the archaeal cell sheath from an amyloid-like protein
Nature Communications (2023)
-
An archaellum filament composed of two alternating subunits
Nature Communications (2022)
-
Cryo-electron microscopy reveals two distinct type IV pili assembled by the same bacterium
Nature Communications (2020)
-
The structures of two archaeal type IV pili illuminate evolutionary relationships
Nature Communications (2020)