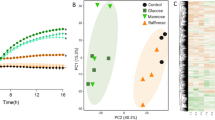
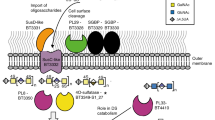
Abstract
Here we employ a ‘systems structural biology’ approach to functionally characterize an unconventional α-glucan metabolic pathway from the food-borne pathogen Listeria monocytogenes (Lm). Crystal structure determination coupled with basic biochemical and biophysical assays allowed for the identification of anabolic, transport, catabolic and regulatory portions of the cycloalternan pathway. These findings provide numerous insights into cycloalternan pathway function and reveal the mechanism of repressor, open reading frame, kinase (ROK) transcription regulators. Moreover, by developing a structural overview we were able to anticipate the cycloalternan pathway's role in the metabolism of partially hydrolysed starch derivatives and demonstrate its involvement in Lm pathogenesis. These findings suggest that the cycloalternan pathway plays a role in interspecies resource competition—potentially within the host gastrointestinal tract—and establish the methodological framework for characterizing bacterial systems of unknown function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
14 July 2017
In the PDF version of this article previously published, the year of publication provided in the footer of each page and in the 'How to cite' section was erroneously given as 2017, it should have been 2016. This error has now been corrected. The HTML version of the article was not affected.
References
Edwards, A. M. et al. Too many roads not taken. Nature 470, 163–165 (2011).
Hermann, J. C. et al. Structure-based activity prediction for an enzyme of unknown function. Nature 448, 775–779 (2007).
Konc, J., Hodošček, M., Ogrizek, M., Trykowska Konc, J. & Janežič, D. Structure-based function prediction of uncharacterized protein using binding sites comparison. PLoS Comput. Biol. 9, e1003341 (2013).
Zhao, S. et al. Discovery of new enzymes and metabolic pathways by using structure and genome context. Nature 502, 698–702 (2013).
Vetting, M. W. et al. Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes. Biochemistry 54, 909–931 (2015).
Anderson, W. F. Structural genomics and drug discovery for infectious diseases. Infect. Disord. Drug Targets 9, 507–517 (2009).
Vazquez-Boland, J. A. et al. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640 (2001).
Renier, S., Micheau, P., Talon, R., Hébraud, M. & Desvaux, M. Subcellular localization of extracytoplasmic proteins in monoderm bacteria: rational secretomics-based strategy for genomic and proteomic analyses. PLoS ONE 7, e42982 (2012).
Suzuki, N. et al. Structural elucidation of the cyclization mechanism of α-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase. J. Biol. Chem. 289, 12040–12051 (2014).
Nishimoto, T. et al. Purification and characterization of glucosyltransferase and glucanotransferase involved in the production of cyclic tetrasaccharide in Bacillus globisporus C11. Biosci. Biotechnol. Biochem. 66, 1806–1818 (2002).
Kim, Y. K., Kitaoka, M., Hayashi, K., Kim, C. H. & Côté, G. L. A synergistic reaction mechanism of a cycloalternan-forming enzyme and a d-glucosyltransferase for the production of cycloalternan in Bacillus sp. NRRL B-21195. Carbohydr. Res. 338, 2213–2220 (2003).
Cote, G. L. & Biely, P. Enzymically produced cyclic α-1,3-linked and α-1,6-linked oligosaccharides of d-glucose. Eur. J. Biochem. 226, 641–648 (1994).
Watanabe, K., Hata, Y., Kizaki, H., Katsube, Y. & Suzuki, Y. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0 Å resolution: structural characterization of proline-substitution sites for protein thermostabilization. J. Mol. Biol. 269, 142–153 (1997).
Kazanov, M. D., Li, X., Gelfand, M. S., Osterman, A. L. & Rodionov, D. A. Functional diversification of ROK-family transcriptional regulators of sugar catabolism in the thermotogae phylum. Nucleic Acids Res. 41, 790–803 (2013).
Wurtzel, O. et al. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 8, 583 (2012).
Schumacher, M. A. et al. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 21, 1210–1218 (2002).
Wong, J. J., Lu, J., Edwards, R. A., Frost, L. S. & Glover, J. N. Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM. Nucleic Acids Res. 39, 6775–6788 (2011).
Bréchemier-Baey, D., Domínguez-Ramírez, L. & Plumbridge, J. The linker sequence, joining the DNA-binding domain of the homologous transcription factors, Mlc and NagC, to the rest of the protein, determines the specificity of their DNA target recognition in Escherichia coli. Mol. Microbiol. 85, 1007–1019 (2012).
Brechemier-Baey, D., Dominguez-Ramirez, L., Oberto, J. & Plumbridge, J. Operator recognition by the ROK transcription factor family members, NagC and Mlc. Nucleic Acids Res. 43, 361–372 (2015).
Gopal, S. et al. Maltose and maltodextrin utilization by Listeria monocytogenes depend on an inducible ABC transporter which is repressed by glucose. PLoS ONE 5, e10349 (2010).
Hibbing, M. E., Fuqua, C., Parsek, M. R. & Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010).
Mashburn, L. M., Jett, A. M., Akins, D. R. & Whiteley, M. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187, 554–566 (2005).
Miethke, M. & Marahiel, M. A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451 (2007).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276, 307–326 (1997).
McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61, 458–464 (2005).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Painter, J. & Merritt, E. A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D 62, 439–450 (2006).
Eschenfeldt, W. H., Lucy, S., Millard, C. S., Joachimiak, A. & Mark, I. D. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115 (2009).
Côté, G. L. & Sheng, S. Penta-, hexa-, and heptasaccharide acceptor products of alternansucrase. Carbohydr. Res. 341, 2066–2072 (2006).
Kelley, L. A. & Sternberg, M. J. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 (2009).
Eschenfeldt, W. H. et al. New LIC vectors for production of proteins from genes containing rare codons. J. Struct. Funct. Genomics 14, 135–144 (2013).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Jones, D. T., Taylor, W. R. & Thornton, J. M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 (1992).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Luan, C. H., Light, S. H., Dunne, S. F. & Anderson, W. F. Ligand screening using fluorescence thermal shift analysis (FTS). Methods Mol. Biol. 1140, 263–289 (2014).
Port, G. C. & Freitag, N. E. Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect. Immun. 75, 5886–5897 (2007).
Bruno, J. C. Jr & Freitag, N. E. Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS ONE 5, e15138 (2010).
Auerbuch, V., Lenz, L. L. & Portnoy, D. A. Development of a competitive index assay to evaluate the virulence of Listeria monocytogenes actA mutants during primary and secondary infection of mice. Infect. Immun. 69, 5953–5957 (2001).
Acknowledgements
The Center for Structural Genomics of Infectious Diseases has been funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Department of Health and Human Services, under contract nos. HHSN272200700058C and HHSN272201200026C (to W.F.A.). This work was supported by NIH grants R01 AI083241 and AI083241-03S1 (to N.E.F.) and F32 AI 115954 (to L.A.C.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research meprogram (grant no. 085P1000817). This work was supported by the Northwestern University High Throughput Analysis Laboratory, the Northwestern University Keck Biophysics Facility and a Cancer Center Support Grant (NCI CA060553). The authors thank C.-H. Luan, G. Minasov, Z. Wawrzak and B. Xayarath for assisting with experiments and S. Almo and G. Côté for providing reagents.
Author information
Authors and Affiliations
Contributions
S.H.L., L.A.C., N.E.F. and W.F.A. designed experiments. S.H.L., L.A.C. and A.S.H. performed experiments. S.H.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Figures 1–10, Supplementary Tables 1–3 and Supplementary References (PDF 1402 kb)
Rights and permissions
About this article
Cite this article
Light, S., Cahoon, L., Halavaty, A. et al. Structure to function of an α-glucan metabolic pathway that promotes Listeria monocytogenes pathogenesis. Nat Microbiol 2, 16202 (2017). https://doi.org/10.1038/nmicrobiol.2016.202
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.202
This article is cited by
-
MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding
Nature Communications (2023)
-
Structure of the transcription open complex of distinct σI factors
Nature Communications (2023)
-
Comparison of carbohydrate ABC importers from Mycobacterium tuberculosis
BMC Genomics (2021)
-
A novel intracellular dextranase derived from Paenibacillus sp. 598K with an ability to degrade cycloisomaltooligosaccharides
Applied Microbiology and Biotechnology (2019)
-
A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria
Nature (2018)