Abstract
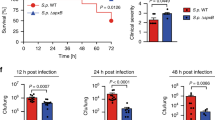
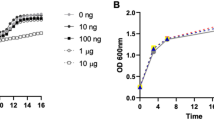
The mechanisms by which pathogens evade elimination without affecting host fitness are not well understood. For the pathogen Pseudomonas aeruginosa, this evasion appears to be triggered by excretion of the quorum-sensing molecule 2-aminoacetophenone, which dampens host immune responses and modulates host metabolism, thereby enabling the bacteria to persist at a high burden level. Here, we examined how 2-aminoacetophenone trains host tissues to become tolerant to a high bacterial burden, without compromising host fitness. We found that 2-aminoacetophenone regulates histone deacetylase 1 expression and activity, resulting in hypo-acetylation of lysine 18 of histone H3 at pro-inflammatory cytokine loci. Specifically, 2-aminoacetophenone induced reprogramming of immune cells occurs via alterations in histone acetylation of immune cytokines in vivo and in vitro. This host epigenetic reprograming, which was maintained for up to 7 days, dampened host responses to subsequent exposure to 2-aminoacetophenone or other unrelated pathogen-associated molecules. The process was found to involve a distinct molecular mechanism of host chromatin regulation. Inhibition of histone deacetylase 1 prevented the immunomodulatory effects of 2-aminoacetophenone. These observations provide the first mechanistic example of a quorum-sensing molecule regulating a host epigenome to enable tolerance of infection. These insights have enormous potential for developing preventive treatments against bacterial infections.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ruby, T. & Monack, D. M. At home with hostility: How do pathogenic bacteria evade mammalian immune surveillance to establish persistent infection? F1000 Biol. Rep. 3, 1 (2011).
Reddick, L. E. & Alto, N. M. Bacteria fighting back: how pathogens target and subvert the host innate immune system. Mol. Cell 54, 321–328 (2014).
Finlay, B. B. & McFadden, G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782 (2006).
Ayres, J. S. & Schneider, D. S. Tolerance of infections. Annu. Rev. Immunol. 30, 271–294 (2012).
Soares, M. P., Gozzelino, R. & Weis, S. Tissue damage control in disease tolerance. Trends Immunol. 35, 483–494 (2014).
Medzhitov, R., Schneider, D. S. & Soares, M. P. Disease tolerance as a defense strategy. Science 335, 936–941 (2012).
Read, A. F., Graham, A. L. & Raberg, L. Animal defenses against infectious agents: is damage control more important than pathogen control. PLoS Biol. 6, e000004 (2008).
Schmid-Hempel, P. Immune defence, parasite evasion strategies and their relevance for ‘macroscopic phenomena’ such as virulence. Phil. Trans. R. Soc. Lond. B 364, 85–98 (2009).
Mehta, S. & Jeffrey, K. L. Beyond receptors and signaling: epigenetic factors in the regulation of innate immunity. Immunol. Cell Biol. 93, 233–244 (2015).
Netea, M. G., Latz, E., Mills, K. H. & O'Neill, L. A. Innate immune memory: a paradigm shift in understanding host defense. Nat. Immunol. 16, 675–679 (2015).
Saeed, S. et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014).
Cheng, S. C. et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014).
Bandyopadhaya, A. et al. The quorum sensing volatile molecule 2-amino acetophenon modulates host immune responses in a manner that promotes life with unwanted guests. PLoS Pathogens 8, e1003024 (2012).
Tzika, A. A. et al. A small volatile bacterial molecule triggers mitochondrial dysfunction in murine skeletal muscle. PLoS ONE 8, e74528 (2013).
Kesarwani, M. et al. A quorum sensing regulated small volatile molecule reduces acute virulence and promotes chronic infection phenotypes. PLoS Pathogens 7, e1002192 (2011).
Cox, C. D. & Parker, J. Use of 2-aminoacetophenone production in identification of Pseudomonas aeruginosa. J. Clin. Microbiol. 9, 479–484 (1979).
Dickschat, J. S., Martens, T., Brinkhoff, T., Simon, M. & Schulz, S. Volatiles released by a Streptomyces species isolated from the North Sea. Chem. Biodivers. 2, 837–865 (2005).
Yamada-Onodera, K., Takase, Y. & Tani, Y. Purification and characterization of 2-aminoacetophenone reductase of newly isolated Burkholderia sp. J. Biosci. Bioeng. 104, 416–419 (2007).
Thiel, V. et al. Identification and biosynthesis of tropone derivatives and sulfur volatiles produced by bacteria of the marine Roseobacter clade. Org. Biomol. Chem. 8, 234–246 (2010).
Scott-Thomas, A. J. et al. 2-Aminoacetophenone as a potential breath biomarker for Pseudomonas aeruginosa in the cystic fibrosis lung. BMC Pulm. Med. 10, 56 (2010).
Rutherford, S. T. & Bassler, B. L. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect Med. 2, a012427 (2012).
Parker, C. T. & Sperandio, V. Cell-to-cell signalling during pathogenesis. Cell Microbiol. 11, 363–369 (2009).
Deziel, E. et al. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-l-homoserine lactones. Mol. Microbiol. 55, 998–1014 (2005).
Xiao, G. et al. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 62, 1689–1699 (2006).
Que, Y. A. et al. A quorum sensing small volatile molecule promotes antibiotic tolerance in bacteria. PLoS ONE 8, e80140 (2013).
Bierne, H., Hamon, M. & Cossart, P. Epigenetics and bacterial infections. Cold Spring Harb. Perspect Med. 2, a010272 (2012).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Hamon, M. A. & Cossart, P. Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe. 4, 100–109 (2008).
Villagra, A., Sotomayor, E. M. & Seto, E. Histone deacetylases and the immunological network: implications in cancer and inflammation. Oncogene 29, 157–173 (2010).
Halili, M. A. et al. Differential effects of selective HDAC inhibitors on macrophage inflammatory responses to the Toll-like receptor 4 agonist LPS. J. Leukoc. Biol. 87, 1103–1114 (2010).
Nishioka, C. et al. MS-275, a novel histone deacetylase inhibitor with selectivity against HDAC1, induces degradation of FLT3 via inhibition of chaperone function of heat shock protein 90 in AML cells. Leuk. Res. 32, 1382–1392 (2008).
Lobera, M. et al. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat. Chem. Biol. 9, 319–325 (2013).
Malvaez, M. et al. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc. Natl Acad. Sci. USA 110, 2647–2652 (2013).
Balasubramanian, S. et al. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 22, 1026–1034 (2008).
Lee, J. H. et al. Development of a histone deacetylase 6 inhibitor and its biological effects. Proc. Natl Acad. Sci. USA 110, 15704–15709 (2013).
Avalos, J. L., Bever, K. M. & Wolberger, C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD+ cosubstrate specificity of a Sir2 enzyme. Mol. Cell 17, 855–868 (2005).
Rahme, L. G. et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902 (1995).
Rahme, L. G. et al. Plants and animals share functionally common bacterial virulence factors. Proc. Natl Acad. Sci. USA 97, 8815–8821 (2000).
Lau, G. W. et al. The Drosophila melanogaster toll pathway participates in resistance to infection by the Gram-negative human pathogen Pseudomonas aeruginosa. Infect. Immun. 71, 4059–4066 (2003).
Mahajan-Miklos, S., Tan, M. W., Rahme, L. G. & Ausubel, F. M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell 96, 47–56 (1999).
Snitkin, E. S. & Segre, J. A. Pseudomonas aeruginosa adaptation to human hosts. Nat. Genet. 47, 2–3 (2015).
Garcia-Garcia, J. C., Barat, N. C., Trembley, S. J. & Dumler, J. S. Epigenetic silencing of host cell defense genes enhances intracellular survival of the Rickettsial pathogen Anaplasma phagocytophilum. PLoS Pathogens 5, e1000488 (2009).
Paschos, K. & Allday, M. J. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol. 18, 439–447 (2010).
Aung, H. T. et al. LPS regulates proinflammatory gene expression in macrophages by altering histone deacetylase expression. FASEB J. 20, 1315–1327 (2006).
Eskandarian, H. A. et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science 341, 1238858 (2013).
Quintin, J., Cheng, S. C., van der Meer, J. W. & Netea, M. G. Innate immune memory: towards a better understanding of host defense mechanisms. Curr. Opin. Immunol. 29, 1–7 (2014).
Li, R. et al. Attenuated Bordetella pertussis protects against highly pathogenic influenza A viruses by dampening the cytokine storm. J. Virol. 84, 7105–7113 (2010).
Khajanchi, B. K., Kirtley, M. L., Brackman, S. M. & Chopra, A. K. Immunomodulatory and protective roles of quorum-sensing signaling molecules N-acyl homoserine lactones during infection of mice with Aeromonas hydrophila. Infect. Immun. 79, 2646–2657 (2011).
Wheeler, D. S. et al. Induction of endotoxin tolerance enhances bacterial clearance and survival in murine polymicrobial sepsis. Shock 30, 267–273 (2008).
Yan, Q. et al. Nuclear factor-κB binding motifs specify Toll-like receptor-induced gene repression through an inducible repressosome. Proc. Natl Acad. Sci. USA 109, 14140–14145 (2012).
Ploeger, D. T. et al. Cell plasticity in wound healing: paracrine factors of M1/ M2 polarized macrophages influence the phenotypical state of dermal fibroblasts. Cell Commun. Signal. 11, 29 (2013).
Shechter, D., Dormann, H. L., Allis, C. D. & Hake, S. B. Extraction, purification and analysis of histones. Nat. Protoc. 2, 1445–1457 (2007).
Acknowledgements
The authors thank S. Mehta and M. Basavappa for their help with the isolation of human primary macrophages and mouse primary macrophages, respectively. The authors thank A. Ballok and A. Power Smith of Write Science Right for editing the manuscript. This work was supported by Shriners grants (nos. 87100 and 85200) and in part by NIH R33AI105902 and Cystic Fibrosis Foundation grant no. CFF#11P0. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
A.B. designed the study and conducted the experiments, performed data analysis and wrote the manuscript. A.T. performed ChIP-PCR experiments and analysis of the data and wrote the manuscript. D.M. contributed to the in vivo experiments. K.L.J. supervised ChIP-PCR experiments. L.G.R. designed the study, supervised the research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
L.G.R. is the scientific founder, consultant and scientific advisory board member of Spero Therapeutics LLC (no funding was received from Spero). The other authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Figures 1–8, Supplementary Table 1 (PDF 2561 kb)
Rights and permissions
About this article
Cite this article
Bandyopadhaya, A., Tsurumi, A., Maura, D. et al. A quorum-sensing signal promotes host tolerance training through HDAC1-mediated epigenetic reprogramming. Nat Microbiol 1, 16174 (2016). https://doi.org/10.1038/nmicrobiol.2016.174
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.174
This article is cited by
-
Tackling recalcitrant Pseudomonas aeruginosa infections in critical illness via anti-virulence monotherapy
Nature Communications (2022)
-
Human M1 macrophages express unique innate immune response genes after mycobacterial infection to defend against tuberculosis
Communications Biology (2022)
-
Epigenetic interaction of microbes with their mammalian hosts
Journal of Biosciences (2021)
-
Heterologous expression of AHL lactonase AiiK by Lactobacillus casei MCJΔ1 with great quorum quenching ability against Aeromonas hydrophila AH-1 and AH-4
Microbial Cell Factories (2020)
-
Whole-organism phenotypic screening for anti-infectives promoting host health
Nature Chemical Biology (2018)