Abstract
During its egress, vaccinia virus transiently recruits AP-2 and clathrin after fusion with the plasma membrane. This recruitment polarizes the viral protein A36 beneath the virus, enhancing actin polymerization and the spread of infection. We now demonstrate that three NPF motifs in the C-terminus of A36 recruit AP-2 and clathrin by interacting directly with the Epsin15 homology domains of Eps15 and intersectin-1. A36 is the first identified viral NPF motif containing protein shown to interact with endocytic machinery. Vaccinia still induces actin tails in the absence of the A36 NPF motifs. Their loss, however, reduces the cell-to-cell spread of vaccinia. This is due to a significant reduction in virus release from infected cells, as the lack of intersectin-1 recruitment leads to a loss of Cdc42 activation, impairing N-WASP-driven Arp2/3-mediated actin polymerization. Our results suggest that initial A36-mediated virus release plays a more important role than A36-driven super-repulsion in promoting the cell-to-cell spread of vaccinia.
Similar content being viewed by others
During vaccinia virus egress, intracellular enveloped virus (IEV) fuses with the plasma membrane to liberate infectious virions into the surrounding environment1. Cell-associated enveloped virions (CEVs) that remain attached to the outside of the cell, however, stimulate Arp2/3 complex-dependent actin polymerization to enhance their spread into neighbouring cells2–5. CEVs stimulate actin polymerization by inducing Src and Abl family kinase-mediated phosphorylation of tyrosine 112 and 132 of the integral viral membrane protein A36 (refs 3,6–9). When phosphorylated, tyrosine 112 recruits Nck, which interacts with a complex of WIP and N-WASP, the latter of which stimulates Arp2/3 complex-mediated actin polymerization3,10–13. Phosphorylation of tyrosine 132 of A36 leads to the recruitment of Grb2, which helps stabilize Nck, WIP and N-WASP to enhance actin polymerization6,12,13. In addition, Cdc42, an activator of N-WASP, also helps facilitate vaccinia actin tail formation14. The ability of Cdc42 to bind N-WASP depends on the RhoGEF intersectin-1, which is recruited beneath CEVs14. Intersectin-1 is a scaffolding protein involved in the initial steps of clathrin-mediated endocytosis that interacts with both Cdc42 and N-WASP15,16. Intersectin-1 also binds to the plasma membrane clathrin adaptor AP-2 (refs 16,17), which is recruited to CEVs, together with clathrin, before actin tail formation18. AP-2 and clathrin do not remain associated with CEV undergoing actin-based motility. Nevertheless, their transient recruitment helps polarize A36 and N-WASP on the virus, resulting in more rapid and sustained actin polymerization18.
We have previously found that intersectin-1 is present on the virus in the absence of AP-2 (ref. 14). The basis of intersectin-1 recruitment and whether it is responsible for localizing AP-2 and clathrin to the virus in addition to activating Cdc42 remains to be established. We now report that A36 contains three NPF motifs that interact with the Eps15 homology (EH) domains in intersectin-1 and Eps15 to promote the release of virus from infected cells and their subsequent spread.
Results
A36 contains three NPF motifs that interact with intersectin-1 and Eps15
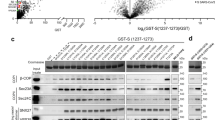
We previously found that endogenous AP-2 associates with the cytoplasmic domain of A36 (residues 24–221)18. This suggests that A36, the viral protein required for vaccinia actin tail formation, may also recruit AP-2 and clathrin beneath CEVs on the plasma membrane. Transient expression of A36 in HeLa cells infected with the Western Reserve (WR) strain of vaccinia virus lacking the endogenous protein (ΔA36R virus) results in its incorporation into virus particles, leading to the formation of actin tails that are 3.0 ± 0.06 µm in length (Fig. 1). Expression of a series of C-terminal deletion mutants reveals that the loss of the last 35 residues of A36 results in significantly longer actin tails (5.8 ± 0.13 µm) (Fig. 1c,d), as previously observed in cells treated with RNAi against AP-2 (ref. 18). Furthermore, GST pulldown assays reveal that A36 deletion mutants inducing long actin tails cannot interact with AP-2 (Fig. 2a). Additional pulldown assays on a series of N-terminal deletions demonstrates that the C terminus of A36 from residue 155 onwards is required to associate with AP-2 (Fig. 2b). This suggests that residues 155–197 of A36 are important for AP-2 binding. However, this region of A36 lacks any canonical AP-2 binding motifs (YxxΦ, D/ExxxL/I, FxNPxY)19. Nevertheless, it does contain three NPF motifs (Fig. 2c). Rather than bind AP-2 directly, NPF motifs interact with EH domains, which are commonly found in endocytic proteins including intersectin-1 and Eps15 (Fig. 2d)19,20. The three A36 NPF motifs are conserved in all sequenced orthopoxvirus genomes, with the exception of the Ectromelia virus A36 homologue (ECTV-Mos-142), which is truncated after residue 161 (Fig. 2c). The actin tail nucleator of Yaba-like disease virus, YL126, and its related homologues in other chordopoxviruses also lack NPF motifs (Fig. 2c).
a, Schematic representation of A36 highlighting its transmembrane domain, kinesin-1 interacting WE/WD motifs and the two tyrosines that, when phosphorylated, bind Nck and Grb2. The A36 C-terminal deletion mutants are indicated. b, Immunofluorescence analysis reveals that expression of A36 in ΔA36R-virus-infected cells rescues the ability of extracellular virus (green) to induce actin tails (red). Scale bars, 10 µm and 2 µm (inset). c, Representative images of actin tails formed by the A36 C-terminal deletion mutants in ΔA36R-virus-infected cells. Scale bar, 2 µm. d, Quantification of the length of actin tails formed by the A36 C-terminal deletion mutants in ΔA36R-virus-infected cells. All immunofluorescence images are representative of three independent experiments, from which a total of 100 tails were measured per condition. Error bars represent s.e.m., and Tukey's multiple comparisons test was used to determine statistical significances. ***P < 0.001.
a, Immunoblot analysis of pulldowns with GST-tagged C-terminal deletions of the A36 cytoplasmic domain reveal that mutants inducing long actin tails do not interact with AP-2. b, Immunoblot analysis of pulldowns with N-terminal deletions of the A36 cytoplasmic domain reveal that residues 155 to 221 are required to interact with AP-2. c, Sequence alignment of A36 homologues from the indicated poxviruses highlighting the three conserved NPF motifs together with YL126 from Yaba-like disease virus. d, Schematic representation of the domain organization of intersectin-1L (ITSN-1L) and Eps15. e, Immunoblot analysis of GFP-trap pulldowns reveals all three A36 NPF motifs are required to associate with AP-2, intersectin-1L (ITSN1) and Eps15. f, GFP-trap pulldowns reveal that the EH domains of intersectin-1L (ITSN1) and Eps15 are required to interact with the cytoplasmic domain of A36. All immunoblots are representative of at least three independent experiments.
By performing GFP-trap pulldown assays, we found that only the cytoplasmic domain of A36 containing three NPF motifs can interact with intersectin-1, Eps15 and AP-2 (Fig. 2e). The EH domains of intersectin-1 and Eps15 are also required for their association with A36 (Fig. 2f). Moreover, a peptide corresponding to residues 158–197 of A36 containing all three NPF motifs binds directly to recombinant EH domains of intersectin-1 and Eps15 in vitro (Fig. 3a,b). No binding was observed when all three NPF motifs are changed to alanine or the amino-acid sequence is scrambled (Fig. 3b). Pulldown assays with peptides containing different combinations of NPF to alanine substitutions demonstrate that loss of the third, but not the first or second NPF motifs, reduces binding to the intersectin-1 and Eps15 EH domains (Fig. 3c). The third NPF motif can also confer binding in the absence of the other two motifs (Fig. 3c). Peptide pulldowns on HeLa cell lysates confirm that the third NPF is sufficient to bind both intersectin-1 and Eps15 (Fig. 3d). Interestingly, loss of the third NPF only abolishes the interaction with intersectin-1. Consistent with this, Eps15, but not intersectin-1, is able to bind to a peptide containing only the second NPF motif (Fig. 3d).
a, Sequence of the A36 peptide (residues 158–197) containing the three NPF motifs (red) used in peptide pulldown assays. Alanine substitutions of the three NPF motifs are indicated together with the sequence of the scrambled control peptide (Scr). b, Coomassie-stained gel demonstrating that the A36 (residues 158–197) but not the scrambled (Scr) or ΔNPF1–3 (Δ1–3) peptides bind directly to recombinant GST-tagged ITSN1 EH1–2 and Eps15 EH1–3. IN and B represent input protein and beads alone, respectively. c, Interaction of GST–ITSN1 EH1–2 and GST–Eps15 EH1–3 with the indicated A36 peptides. IN and B represent input protein and beads alone, respectively. d, Immunoblot analysis of peptide pulldown assays on HeLa cell lysates. e, Images show that endogenous intersectin-1 and Eps15 are recruited to CEVs (indicated by white arrows). Scale bar, 10 µm. f, Stills from live cell imaging (Supplementary Video 1) reveals intersectin-1 and Eps15 are both recruited to CEVs inducing actin tails (indicated by yellow arrows). Scale bar, 10 µm. g, Immunoblot analysis of GFP-trap pulldowns on lysates from siRNA-treated HeLa cells reveals that the interaction of AP-2 with the cytoplasmic domain of A36 requires Eps15 and intersectin-1.
In agreement with our pulldown assays, immunofluorescence analysis reveals that CEVs with or without actin tails recruit both intersectin-1 and Eps15 (Fig. 3e,f and Supplementary Video 1). This suggests that both proteins may be working as a complex to recruit AP-2. Consistent with this, GFP-trap pulldowns on lysates from ΔA36R-infected cells treated with intersectin-1 or Eps15 siRNA demonstrate that the interaction of A36 with AP-2 is impaired in the absence of either protein (Fig. 3g). Furthermore, loss of either intersectin-1 or Eps15 leads to similarly long actin tails (Supplementary Fig. 1a,b). The A36 NPFs appear to be specific for the EH domains of intersectin-1 and Eps15 as the virus did not recruit C-terminal EH domain proteins 1–4 (EHD1–4) (Supplementary Fig. 1c,d). Our observations suggest that the A36 NPF motifs recruit AP-2 and clathrin by interacting directly with a complex of intersectin-1 and Eps15.
A36 NPF motifs recruit intersectin-1, Eps15, AP-2 and clathrin to CEVs
To directly assess the role of Eps15 and intersectin-1 in AP-2:clathrin recruitment, we generated recombinant viruses in which the individual NPF motifs were mutated to alanine (ΔNPF1, ΔNPF2 and ΔNPF3). We also generated a virus expressing A36 lacking all three NPF motifs (ΔNPF1–3) and a truncated version of the protein (residues 1–155). The different A36 mutants were expressed to the same extent and with similar kinetics as the wild-type protein (Fig. 4a). In infected HeLa cells, AP-2 and clathrin are transiently recruited to CEV prior to actin tail formation18. In the absence of actin tail formation in Nck and N-WASP deficient MEFs, the frequency of co-localization of AP-2 and clathrin with CEV significantly increases18. We took advantage of this increase to assess the role of the individual NPFs in recruiting AP-2 and clathrin by infecting N-WASP −/− MEFs. We found that the single A36 ΔNPF mutant viruses recruit intersectin-1, Eps15, AP-2 and clathrin to the same extent as WR (Fig. 4b). In contrast, the ΔNPF1–3 and A36 1–155 viruses had a dramatic decrease in intersectin-1, Eps15, AP-2 and clathrin recruitment. Live cell imaging confirmed that GFP-tagged intersectin-1 and Eps15 are no longer recruited to the ΔNPF1-3 virus inducing actin tails (Supplementary Fig. 2a and Supplementary Videos 2 and 3). Vaccinia expressing YL126 of Yaba-like disease virus, which lacks NPF motifs, in place of A36, is also deficient in intersectin-1, Eps15, AP-2 and clathrin recruitment (Supplementary Fig. 2b). Our observations clearly demonstrate that the three A36 NPF motifs are necessary and sufficient to recruit intersectin-1, Eps15, AP-2 and clathrin to CEVs.
a, Immunoblot analysis of A36 expression during infection with the indicated viruses. The time (h.p.i., hours post-infection) is indicated, and vinculin represents a loading control. b, Representative immunofluorescence images from three independent experiments showing that clathrin, AP-2, intersectin-1 and Eps15 are recruited to WR (white arrows) but not the A36 ΔNPF1–3 or 1–155 viruses in N-WASP −/− MEFs. Scale bar, 2 µm. Graphs show the quantification of the recruitment of clathrin, AP-2, intersectin-1 and Eps15 to WR and the A36 recombinant viruses, where n = 30 cells. Error bars represent s.e.m. from three independent experiments in which a total of 3,000 viruses were analysed. Tukey's multiple comparisons test was used to determine statistical significances. ***P < 0.001; NS, not significant.
NPF motifs promote viral release and spread
We found that mutation of a single NPF motif results in the formation of longer actin tails (Fig. 5a,b). The actin tails induced by the ΔNPF1–3 and A36 1–155 viruses were, however, even longer than the single NPF mutants (Fig. 5a,b). Additionally, only the loss of all three NPF motifs resulted in a faster velocity of the virus (Fig. 5b and Supplementary Video 4). This mirrors the phenotype induced by the depletion of AP-2 and clathrin18. The absolute number of actin tails induced by the NPF mutant viruses did not change compared to WR (Fig. 5b). However, live cell imaging reveals that it takes approximately twice as long for the ΔNPF1–3 virus to induce an actin tail compared to WR (Fig. 5c and Supplementary Fig. 3).
a, Images of actin tails (red) induced by the indicated virus (green) in HeLa cells representative from three independent experiments. Scale bar, 10 µm. b, Quantification of number of actin tails induced per cell, their length and speed. Error bars represent s.e.m. from three independent experiments in which a total of n = 30 cells were analysed for tail number, and n = 1,200 and n = 75 actin tails were measured for length and speed, respectively. See, also, Supplementary Video 4. c, Quantification of the time taken for virus to induce actin tails. Error bars represent s.e.m. from n = 60 tails in three independent experiments. d, Representative images and quantification of plaque sizes produced by the indicated virus in BS-C-1 cells 48 h post-infection (n = 60 plaques in three independent experiments). Scale bar, 5 mm. Tukey's multiple comparisons test was used to determine statistical significances; ***P < 0.001; ****P < 0.0001; NS, not significant.
The ability of vaccinia to undergo A36-dependent actin-driven super-repulsion on recently infected cells enhances viral spread in confluent cell monolayers21. To investigate the possible role of the A36 NPF motifs and clathrin recruitment in super-repulsion, we first examined the size of plaques formed by the different viruses in BS-C-1 cell monolayers. There was no difference in the size of plaques formed by the individual NPF mutants compared to those produced by WR (Fig. 5d). In contrast, both the ΔNPF1–3 and A36 1–155 viruses induce significantly smaller plaques, consistent with the possible involvement of A36-mediated recruitment of clathrin in promoting viral spread by super repulsion. To confirm if this is the case, we generated recombinant viruses in which the expression of A36 and its ΔNPF1–3 mutant are under control of a late 4b promoter. Immunoblot analysis confirmed that expression of A36late and ΔNPF1–3late is only observed from 6 h post-infection and is blocked by AraC, an inhibitor of late viral protein expression (Supplementary Fig. 4b). As seen previously21, late expression of A36 results in an ∼50% decrease in plaque size compared to those formed by WR (Fig. 6a). A similar decrease is observed when A36 ΔNPF1–3 is expressed from its endogenous early promoter (Fig. 6a). These data suggest that the NPF motifs enhance viral spread by facilitating super-repulsion of the virus on recently infected cells. The late expression of ΔNPF1–3late, however, did not lead to any further reduction in plaque size (Fig. 6a). Given that A36-dependent actin polymerization also promotes virus release22, we wondered whether the reduced spread of the ΔNPF1–3 virus also involves impaired viral release from infected cells. Titering infectious virus in the cell media, we found that loss of the three A36 NPF motifs leads to a significant drop in virus release (Fig. 6b). The levels of total virus produced during infection, however, did not change (Supplementary Fig. 4a). Furthermore, a similar reduction of viral release was also observed with A36late (Fig. 6b). This suggests that the A36 NPF motifs enhance viral spread primarily by promoting viral release rather than facilitating super-repulsion.
a, Representative images and quantification of plaque sizes produced in BS-C-1 cells 48 h post-infection with the indicated viruses (n = 75 plaques in three independent experiments). Scale bar, 5 mm. b, Quantification of extracellular enveloped virus (EEV) release at 16 h post-infection. c, Representative plaques formed by WR and the A36 ΔNPF1–3 virus 48 h post-infection in N-WASP −/− MEFs expressing GFP–N-WASP or GFP–N-WASP–H208D (n = 150 plaques in three independent experiments). Scale bar, 5 mm. d, Quantification of EEV release from GFP–N-WASP MEFs at 16 h post-infection. e, Quantification of actin tail lengths induced by WR and the A36 ΔNPF1–3 virus in cells expressing intersectin-1L, intersectin-1S (GEF deficient) or treated with ZCL278. Error bars represent s.e.m. from three independent experiments in which a total of 300 tails were measured in each condition. f, Representative images and quantification of actin tail length induced by the indicated viruses in cells expressing GFP-tagged Cdc42 or its constitutively active mutant (Cdc42–Q61L). Scale bar, 5 µm. Error bars represent s.e.m. from three independent experiments in which a total of 300 tails were measured in each condition. g, Representative immunofluorescent plaque images together with quantification of their size and number in +/− or −/− Cdc42 fibroblastoid cells 105 h post-infection with the indicated viruses. Scale bar, 5 mm. Error bars represent s.e.m. from three independent experiments in which 60, 75, 25 and 35 plaques were analysed, respectively. In all panels, error bars represent s.e.m. over three independent experiments. Tukey's multiple comparisons test was used to determine statistical significances. **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant.
Intersectin-1-mediated activation of Cdc42 enhances virus release
Given this, we investigated whether intersectin-1 enhances viral release by promoting Cdc42-mediated activation of N-WASP and/or recruiting clathrin via AP-2. To distinguish between these possibilities, we examined the size of plaques formed on N-WASP −/− MEFs stably expressing GFP–N-WASP–H208D, which cannot interact with Cdc42 (Supplementary Fig. 4c)23. We found WR produced larger plaques than the ΔNPF1–3 virus on N-WASP −/− MEFs stably expressing GFP–N-WASP (Fig. 6c). In contrast, both viruses formed similar-sized smaller plaques on cells expressing N-WASP–H208D (Fig. 6c). In the case of WR, we found that significantly less infectious virus was released into the media from N-WASP −/− MEFs expressing GFP-tagged N-WASP–H208D than N-WASP (Fig. 6d). In contrast, the ΔNPF1–3 virus was equally deficient in virus release regardless of the ability of N-WASP to interact with Cdc42 or not. This suggests that NPF-mediated recruitment of intersectin-1 enhances viral spread by promoting the release of vaccinia from infected cells by activating Cdc42. To further explore this possibility, we examined the length of actin tails in cells expressing intersectin-1S, a naturally occurring splice variant that lacks GEF activity (Supplementary Fig. 4d)15. We found that intersectin-1S but not the full-length protein, increases the length of WR actin tails (Fig. 6e and Supplementary Fig. 4d). In contrast, the ΔNPF1–3 virus induces equally long tails regardless of the presence or absence of intersectin-1 GEF activity. The treatment of WR but not ΔNPF1–3 virus-infected cells with ZCL278, a small molecule that inhibits Cdc42–intersectin-1 interactions24, also results in longer actin tails (Fig. 6e and Supplementary Fig. 4e). Expression of Cdc42 or its activated Q61L mutant did not revert the long tail phenotype of the ΔNPF1–3 virus, presumably because both proteins are only weakly recruited to the virus (Fig. 6f). Finally, we examined the impact of loss of Cdc42 by infecting fibroblastoid cells derived from mice lacking Cdc42 (Supplementary Fig. 4f)25. We found that loss of the NPF motifs results in a significant reduction in the size but not number of plaques formed on fibroblastoid cells expressing Cdc42 (Fig. 6g). As expected from its role during virus entry26, loss of Cdc42 results in significantly smaller plaques (Fig. 6g). The size and number of these plaques, however, were not affected by the presence or absence of the A36 NPF motifs (Fig. 6g). Taken together, our data demonstrate that the A36 NPF motifs promote the release and spread of the virus by recruiting intersectin-1, so it can activate Cdc42, allowing it to bind N-WASP to stimulate Arp2/3-driven actin polymerization.
Discussion
A36 contains EH-domain-binding NPF motifs
We have now demonstrated that AP-2 and clathrin recruitment to CEVs depends on three NPF motifs in the integral viral membrane protein A36. NPF motifs interact with EH domains found in proteins primarily involved in endocytosis and vesicle trafficking27,28 NPF motif-containing proteins are found widely, from humans to yeast28,29, making A36 the first example of a viral protein containing functional NPF motifs. The A36 NPF motifs appear to be specific for intersectin-1 and Eps15, as they cannot recruit EHD1–4 proteins to the virus, consistent with the absence of acidic residues downstream of the NPF motifs20. The three NPF motifs are highly conserved in other orthopoxvirus A36 homologues, suggesting they play an important role in promoting viral spread. Despite their strong conservation, there are exceptions, such as the Ectromelia virus actin tail nucleator ECTV-Mos-142, which has 92% sequence homology to the first 157 residues of A36 but lacks the C-terminal region containing the NPF motifs30. An inability of ECTV-Mos-142 to recruit AP-2 via intersectin-1 and Eps15 may account for the reduced robustness of Ectromelia actin tails compared to those nucleated by vaccinia, as in the latter case, recruitment of clathrin enhances their persistence18. Yatapoxvirus actin tail nucleators such as YL126 from Yaba-like disease virus lacking NPF motifs also do not induce robust actin tails31.
Intersectin-1 and Eps15 cooperate to recruit AP-2 and clathrin
As commonly seen with other NPF-containing proteins, A36 contains a tandem arrangement of closely spaced NPF motifs. The moderate affinity of NPF motifs for EH domains, which are also often present in multiple copies, is common for weak cooperative interactions, such as those found during endocytosis27,29,32. Consistent with this, mutation of a single NPF motif is sufficient to abrogate the interaction of intersectin-1 and Eps15 with the isolated cytoplasmic domain of A36 in cell lysates. In contrast, mutation of a single NPF motif in an A36 peptide containing all three motifs does not always abrogate the interaction with intersectin-1 or Eps15 in in vitro pulldown assays. We believe this difference reflects the high concentration of the A36 peptide on the bead, thereby enhancing weak cooperative interactions. A similar situation also appears to occur on the virus, as mutation of a single A36 NPF motif does not result in a loss of intersectin-1, Eps15, AP-2 or clathrin recruitment. Notwithstanding this, our in vitro peptide pulldown assays demonstrate that the three NPF motifs do not have equivalent interactions. In particular, the most C-terminal third NPF appears to play the dominant role in binding the EH domains of intersectin-1 or Eps15. The third A36 NPF motif is also necessary and sufficient to interact with intersectin-1. In contrast, the presence of the second or third but not the first NPF motif is capable of mediating the interaction with Eps15 and AP-2. Our peptide pulldowns on HeLa cell lysates offer an explanation as to how mutation of a single NPF, including the third motif, is not sufficient to abolish recruitment of AP-2 and clathrin to the virus. Moreover, in the context of the virus, the absence of the third A36 NPF motif does not result in loss of intersectin-1 recruitment. Intersectin-1 and Eps15 are both capable of binding directly to AP-2 (refs 17,33). However, they have also been shown to hetero-dimerize to recruit AP-2 (refs 34,35). The presence of intersectin-1 and Eps15 on the same virus particles suggests that both proteins are acting together to recruit AP-2 and clathrin. Consistent with this, RNAi-mediated loss of intersectin-1 or Eps15 is sufficient to induce the formation of long actin tails. Our previous observations also demonstrate that in the absence of AP-2 and clathrin recruitment, vaccinia induces longer actin tails18. This increase in length is the consequence of changes in the polarization of A36 molecules beneath CEVs, which presumably impacts on the number and organization of actin filaments nucleated by the virus. Interestingly, loss of a single NPF motif also results in longer actin tails, even though it does not impact on the ability of the virus to recruit AP-2, clathrin, intersectin-1 or Eps15. This suggests that the organization of A36 beneath CEVs and/or the associated signalling network stimulating Arp2/3-mediated actin polymerization is altered. Indeed, the SH3 domains of intersectin-1 interact directly with both WIP and N-WASP, essential components of the vaccinia signalling network, while its DH-PH domains activate Cdc42 so it can bind N-WASP to promote actin tail formation13–15,36. Understanding how intersectin-1, Eps15, AP-2 and clathrin affect the organization and polarization of A36 and its associated signalling network will require ultrastructural analysis of CEVs at the plasma membrane. Such analysis would provide important information on the nature of the clathrin lattice and whether it varies between individual CEVs as well as its relationship to the actin cytoskeleton.
A36 NPF motifs promote spread of vaccinia by enhancing viral release
The loss of the three NPF motifs and clathrin recruitment does not ultimately impact the ability of the virus to induce actin tails. This is presumably because the density of A36 beneath the CEVs after fusion of the virus with the plasma membrane overcomes an absolute necessity for clathrin-mediated clustering of A36 to stimulate Arp2/3-mediated actin polymerization18,22. Nevertheless, loss of the three A36 NPF motifs does result in smaller plaques, which is indicative of reduced cell-to-cell spread of the virus. The ability of vaccinia to undergo actin-dependent super-repulsion on recently infected cells is thought to provide a major driving force enhancing viral spread in confluent cell monolayers21. It is possible that when an extracellular virus lands on a recently infected cell, NPF-mediated recruitment of clathrin promotes clustering of A36 present in the plasma membrane several hours before new virions are assembled. Clathrin-mediated clustering of A36 would presumably help assemble a more robust signalling platform, making it easier for the virus to initiate and maintain actin polymerization, which is a prerequisite for super-repulsion to occur. Such a scenario has strong parallels with the steps involved in the initiation of receptor-dependent clathrin-mediated endocytosis37,38. While still a possibility, our observations demonstrate that the NPF motifs principally promote viral spread by enhancing the release of virus from infected cells rather than facilitating super-repulsion. They achieve this by recruiting intersectin-1 and activating Cdc42 to drive Arp2/3-mediated actin polymerization through its interaction with N-WASP. In summary, our study has now provided important molecular insights into the recruitment of clathrin and regulation of actin polymerization during virus release.
Methods
Antibodies, drugs, expression clones and cells
The following primary antibodies were used for immunofluorescence and immunoblotting: AP-2 (Abcam, 2730), CHC (Abcam), intersectin-1 (clone 29, BD Biosciences), Eps15 C-20 (Santa Cruz, sc-534), EHD1 (Novus, NBP1-95580), EHD2 (Abcam, ab23935), EHD4 (Novus, H00030844-M01), B5 (ref. 39), A36 (ref. 40), GFP 3E1 monoclonal (Cancer Research UK), GST (G7781), actin AC-74 (Sigma-Aldrich) and Vinculin hVIN-1 (Sigma-Aldrich). Secondary antibodies and Phalloidin Texas Red were obtained from Invitrogen. ZCL278, which inhibits Cdc42–intersectin-1 interactions24, was obtained from Tocris Bioscience and added to a final concentration of 50 µM 4 h post-infection before IEV assembly and actin tail formation.
pE/L expression vectors encoding GFP-tagged Cdc42, Cdc42–QL61, intersectin-1 L and intersectin-1S have been described previously10,14. CMV-based expression vectors for GFP-tagged C-terminal Eps15 homology domain proteins 1–4 (EHD1–4)41–44 were provided by Steve Caplan (University of Nebraska Medical Center) and transfected into HeLa cells 16 h before infection according to refs 45 and 46.
HeLa and BS-C-1 cell lines were maintained in modified Eagle's medium (MEM) supplemented with 10% fetal bovine serum (FBS), 100 units ml–1 penicillin and 100 µg ml–1 streptomycin at 37 °C and 5% CO2. HeLa and BS-C-1 cells are originally from ATCC and the former were obtained from G. Griffiths (Cell Biology programme, EMBL). N-WASP −/− MEFs47 and Cdc42-deficient fibroblastoid cells25 were obtained from Scott Snapper (Boston Children's Hospital, Harvard Medical School) and Cord Brakebusch (University of Copenhagen), respectively and maintained in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 100 units ml−1 penicillin and 100 µg ml–1 streptomycin. All cell lines used in this study have been tested for mycoplasma by the London Research Institute, Cell Services, and no cell line was found in the database of commonly misidentified cell lines (maintained by ICLAC and NCBI Biosample). All cell lines were authenticated by STR profiling by Cell Services at our institute. N-WASP −/− MEFs stably expressing GFP–N-WASP and GFP–N-WASP–H208D as well as HeLa LifeAct-Cherry cells have been previously described12,14,18. The pLVX–Lifeact–iRFP670 lentiviral expression vector was created by inserting iRFP67048 tagged at its N terminus with LifeAct into the Xho1 and BamH1 sites of the pLVX-IRES hygro vector (Clonetech) as previously described49. Lentivirus was prepared and used to infect HeLa cells as described previously50. HeLa cells stably expressing LifeAct–iRFP670 were selected and maintained in culture medium containing 50 µg ml–1 hygromycin B.
Construction of recombinant A36 ΔNPF and late viruses
Recombinant mutant viruses were generated in vaccinia virus strain WR as described previously6. In short, 324 bp upstream and downstream of the A36R genome was amplified from viral genomic DNA and subcloned into the Not1–EcoR1 sites of pBS SKII. A36 NPF–AAA mutants were then generated using two sequential rounds of mutagenesis. HeLa cells infected with WR–ΔA36R were subsequently transfected with the relevant pBS SKII A36R–ΔNPF targeting vectors using Lipofectamine2000 (Invitrogen) as described by the manufacturer. Recombinant A36R–ΔNPF viruses were subsequently isolated based on plaque size over four rounds of plaque purification. Successful recombination at the correct locus was verified by sequencing and by immunoblot analysis. A36 and A36–ΔNPF mutants under late expression were generated by ligation of the 4b promoter (GGATCCATCACGCTTTCGAGTAAAAACTACGAATATAAATAATGCTCGAG) to the 5′ of the A36R open reading frame. The resulting construct was subcloned into the BamH1–Not1 sites of pJS4 plasmid51, which are flanked by the left and right arms of homology to the TK locus in WR. Recombinant viruses were generated and selected for as described above. Late expression of A36 was confirmed by immunoblot analysis and AraC treatment (50 µM). All recombinant viruses were purified through a sucrose cushion before use and storage.
Infection, RNAi treatment and immunofluorescence analysis
Cells were infected 24 h after plating on fibronectin with virus at a multiplicity of infection (MOI) of 2 and fixed at 9 h post-infection to be processed for immunofluoresence as previously described45,46. Fixed cells were imaged with a ×63/1.4 Plan Achromat objective on Zeiss Axioplan2 microscope equipped with a Photometrics Cool Snap HQ cooled charge-coupled device camera. The system was controlled with MetaMorph 6.3r7 software. Where required cells were transfected with 20 µM siRNA as per the Hiperfect fast forward protocol (QIAGEN) and incubated for 48 h before infection for 8 h followed by processing for immunofluorescence analysis or for immunoblot analysis. siRNA oligos targeting AP-2 (AGCAUGUGCACGCUGGCCA), intersectin-1 (GGCCAUAACUGUAGAGGAA) and Eps15 (ATGCTGTAGGTTGAACCATTA) were obtained from (Sigma-Aldrich).
Live cell imaging
HeLa LifeAct cells were plated onto Matek dishes coated with fibronectin one day before infection. Cells were infected with WR and A36 ΔNPF viruses at an MOI of 2 and imaged at 37 °C, in phenol red free MEM with 40 mM HEPES from 7 h post-infection on a Zeiss Axio Observer microscope equipped with a Plan Achromat ×63/1.4 Ph3 M27 oil lens, an Evolve 512 camera and a Yokagawa CSUX spinning disk under the control of Slidebook 6.0 (Intelligent imaging). Where indicated, full-length human Eps15 (Addgene) was cloned into the Not1-BamH1 sites of pE/L RFP expression vector and expressed during infection alongside pE/L GFP–intersectin-1 as previously described14. Videos were analysed for actin tail speed and time from viral fusion to actin tail formation using Metamorph and ImageJ software, respectively, from three independent experiments.
Pulldown assays
To investigate the binding of AP-2 to A36, GST-tagged N- and C-terminal truncations of the cytoplasmic domain of A36 were cloned into the pE/L expression vector3. HeLa cells were transfected with pE/L expression vectors 1 h after infection with ΔA36R; 14 h later, cells were lysed and incubated with glutathione beads. The A36 NPF mutants as well as intersectin-1 and Eps15 lacking their EH domains were cloned into pE/L GFP expression vector and expressed as above. GFP-trap A resin (Chromotek) was used to pull down GFP constructs from HeLa cell lysates and probed for binding. N-terminally biotinylated A36 peptides were synthesized in house and coupled to Streptavidin M-280 Dynabeads beads (ThermoFisher Scientific). Recombinant GST-EH domains of Eps15 (EH1–3) and ITSN1 (EH1–2) were amplified by PCR, cloned into pMW172 vector and expressed in BL21 (DE3) Rosetta cells and purified as previously described52.
Plaque assays and virus release assays
Plaque assays were performed in BS-C-1 cells and N-WASP/Cdc42-deficient cell lines for 48 h or 105 h, respectively, before fixation, and subsequently visualized with crystal violet cell stain or immunofluorescent staining of viral B5 as previously described18. Plaque diameter was quantified manually using ImageJ (NIH). To quantify EEV release, 12-well dishes were seeded with BS-C-1 cells and infected at an MOI of 0.1 in serum-free MEM. Supernatants were collected at 16 h post-infection and titrated in tenfold dilutions on BS-C-1 cells as described above. At 48 h post-infection, cells were stained with crystal violet and the number of EEV released was quantified. All release assays were performed in triplicate and over three independent experiments.
Quantification and figure preparation
Quantification of the number of actin tails per cell was determined in ten randomly selected cells in three independent experiments. Actin tail length was measured using ImageJ for 20 actin tails in 20 individual cells in three independent experiments. The velocity of actin tails was measured using manual tracking software in MetaMorph (Molecular Devices Corporation). HeLa Lifeact-Cherry cells were infected, and three-minute movies of actin tails were acquired at 8 h.p.i. For actin tail speed, 75 virons were quantified in each condition in three independent experiments. Co-localization analysis of endocytic machinery was quantified for 100 randomly selected extracellular virus particles per cell18. All data are presented as means ± s.e.m. and were analysed by one-way analysis of variance with Tukey's multiple comparison test using Prism 6 (GraphPad Software). All figures and graphs were prepared using Prism and Illustrator software.
References
Leite, F. & Way, M. The role of signalling and the cytoskeleton during vaccinia virus egress. Virus Res. 209, 87–99 (2015).
Cudmore, S., Cossart, P., Griffiths, G. & Way, M. Actin-based motility of vaccinia virus. Nature 378, 636–638 (1995).
Frischknecht, F. et al. Actin based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401, 926–929 (1999).
Hollinshead, M. et al. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154, 389–402 (2001).
Ward, B. M. & Moss, B. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75, 11651–11663 (2001).
Scaplehorn, N. et al. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr. Biol. 12, 740–745 (2002).
Newsome, T. P., Scaplehorn, N. & Way, M. SRC mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science 306, 124–129 (2004).
Reeves, P. M. et al. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nature Med. 11, 731–739 (2005).
Newsome, T. P., Weisswange, I., Frischknecht, F. & Way, M. Abl collaborates with Src family kinases to stimulate actin-based motility of vaccinia virus. Cell Microbiol. 8, 233–241 (2006).
Moreau, V. et al. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nature Cell Biol. 2, 441–448 (2000).
Zettl, M. & Way, M. The WH1 and EVH1 domains of WASP and Ena/VASP family members bind distinct sequence motifs. Curr. Biol. 12, 1617–1622 (2002).
Weisswange, I., Newsome, T. P., Schleich, S. & Way, M. The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature 458, 87–91 (2009).
Donnelly, S. K., Weisswange, I., Zettl, M. & Way, M. WIP provides an essential link between Nck and N-WASP during Arp2/3-dependent actin polymerization. Curr. Biol. 23, 999–1006 (2013).
Humphries, A. C., Donnelly, S. K. & Way, M. Cdc42 and the Rho GEF intersectin-1 collaborate with Nck to promote N-WASP-dependent actin polymerisation. J. Cell Sci. 127, 673–685 (2014).
Hussain, N. K. et al. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nature Cell Biol. 3, 927–932 (2001).
Hunter, M. P., Russo, A. & O'Bryan, J. P. Emerging roles for intersectin (ITSN) in regulating signaling and disease pathways. Int. J. Mol. Sci. 14, 7829–7852 (2013).
Pechstein, A. et al. Regulation of synaptic vesicle recycling by complex formation between intersectin 1 and the clathrin adaptor complex AP2. Proc. Natl Acad. Sci. USA 107, 4206–4211 (2010).
Humphries, A. C. et al. Clathrin potentiates vaccinia-induced actin polymerization to facilitate viral spread. Cell Host Microbe 12, 346–359 (2012).
Traub, L. M. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nature Rev. Mol. Cell Biol. 10, 583–596 (2009).
Naslavsky, N. & Caplan, S. EHD proteins key conductors of endocytic transport. Trends Cell Biol. 21, 122–131 (2011).
Doceul, V., Hollinshead, M., van der Linden, L. & Smith, G. L. Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science 327, 873–876 (2010).
Horsington, J. et al. A36-dependent actin filament nucleation promotes release of vaccinia virus. PLoS Pathogens 9, e1003239 (2013).
Miki, H., Sasaki, T., Takai, Y. & Takenawa, T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature 391, 93–96 (1998).
Friesland, A. et al. Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc. Natl Acad. Sci. USA 110, 1261–1266 (2013).
Czuchra, A. et al. Cdc42 is not essential for filopodium formation, directed migration, cell polarization, and mitosis in fibroblastoid cells. Mol. Biol. Cell 16, 4473–4484 (2005).
Mercer, J. et al. Vaccinia virus strains use distinct forms of macropinocytosis for host-cell entry. Proc. Natl Acad. Sci. USA 107, 9346–9351 (2010).
De Beer, T., Carter, R. E., Lobel-Rice, K. E., Sorkin, A. & Overduin, M. Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain. Science 281, 1357–1360 (1998).
Confalonieri, S. & Di Fiore, P. P. The Eps15 homology (EH) domain. FEBS Lett. 513, 24–29 (2002).
Paoluzi, S. et al. Recognition specificity of individual EH domains of mammals and yeast. EMBO J. 17, 6541–6550 (1998).
Lynn, H. et al. Loss of cytoskeletal transport during egress critically attenuates ectromelia virus infection in vivo. J. Virol. 86, 7427–7443 (2012).
Dodding, M. P. & Way, M. Nck- and N-WASP-dependent actin-based motility is conserved in divergent vertebrate poxviruses. Cell Host Microbe 6, 536–550 (2009).
Yamabhai, M. et al. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J. Biol. Chem. 273, 31401–31407 (1998).
Benmerah, A., Begue, B., Dautry-Varsat, A. & Cerf-Bensussan, N. The ear of α-adaptin interacts with the COOH-terminal domain of the Eps 15 protein. J. Biol. Chem. 271, 12111–12116 (1996).
Sengar, A. S., Wang, W., Bishay, J., Cohen, S. & Egan, S. E. The EH and SH3 domain Ese proteins regulate endocytosis by linking to dynamin and Eps15. EMBO J. 18, 1159–1171 (1999).
Wong, K. A. et al. Intersectin (ITSN) family of scaffolds function as molecular hubs in protein interaction networks. PLoS ONE 7, e36023 (2012).
Gryaznova, T. et al. Intersectin adaptor proteins are associated with actin-regulating protein WIP in invadopodia. Cell Signal 27, 1499–1508 (2015).
Humphries, A. C. & Way, M. The non-canonical roles of clathrin and actin in pathogen internalization, egress and spread. Nature Rev. Microbiol. 11, 551–560 (2013).
McMahon, H. T. & Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nature Rev. Mol. Cell Biol. 12, 517–533 (2011).
Hiller, G. & Weber, K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J. Virol. 55, 651–659 (1985).
Röttger, S., Frischknecht, F., Reckmann, I., Smith, G. L. & Way, M. Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J. Virol. 73, 2863–2875 (1999).
Caplan, S. et al. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 21, 2557–2567 (2002)
Naslavsky, N., Rahajeng, J., Sharma, M., Jovic, M. & Caplan, S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol. Biol. Cell 17, 163–177 (2006).
Sharma, M., Naslavsky, N. & Caplan, S. A role for EHD4 in the regulation of early endosomal transport. Traffic 9, 995–1018 (2008).
Simone, L. C., Caplan, S. & Naslavsky, N. Role of phosphatidylinositol 4,5-bisphosphate in regulating EHD2 plasma membrane localization. PLoS ONE 8, e74519 (2013).
Arakawa, Y., Cordeiro, J. V., Schleich, S., Newsome, T. & Way, M. The release of vaccinia virus from infected cells requires RhoA-mDia modulation of cortical actin. Cell Host Microbe 1, 227–240 (2007).
Arakawa, Y., Cordeiro, J. V. & Way, M. F11L-mediated inhibition of RhoA-mDia signaling stimulates microtubule dynamics during vaccinia virus infection. Cell Host Microbe 1, 213–226 (2007).
Snapper, S. B. et al. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nature Cell Biol. 3, 897–904 (2001).
Shcherbakova, D. M. & Verkhusha, V. V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nature Methods 10, 751–754 (2013).
Abella, J. V. et al. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nature Cell Biol. 18, 76–86 (2016).
Barry, D. J., Durkin, C. H., Abella, J. V. & Way, M. Open source software for quantification of cell migration, protrusions, and fluorescence intensities. J. Cell Biol. 209, 163–180 (2015).
Chakrabarti, S., Sisler, J. R. & Moss, B. Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques 23, 1094–1097 (1997).
Boëda, B. et al. Tes, a specific Mena interacting partner, breaks the rules for EVH1 binding. Mol. Cell 28, 1071–1082 (2007).
Acknowledgements
The authors thank S. Snapper (Harvard Medical School) and C. Brakebusch (University of Copenhagen) for providing N-WASP and Cdc42 deficient cells, respectively. The authors also thank N. O'Reilly (Francis Crick Institute) for synthesizing peptides, M. Matsuda (Kyoto University) for the iRFP670 clone and S. Caplan (University of Nebraska Medical Center) for GFP-tagged EHD1–4 clones. The authors acknowledge members of the Way Laboratory and H. Walden (University of Dundee) and D. Stephens (University of Bristol) for comments on the manuscript. Research was supported by Cancer Research UK and the Francis Crick Institute.
Author information
Authors and Affiliations
Contributions
X.S. and M.W. designed the study and wrote the manuscript. X.S. performed and analysed the experiments. I.W. constructed and analysed A36 C-terminal deletion mutants. J.P. generated pLVX-Lifeact-iRFP670 HeLa cells and A.C.H. generated pE/L-GFP-intersectin-1 clones and provided valuable discussions. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-5, Supplementary Video 1-4 legends (PDF 232610 kb)
Supplementary Video 1
Intersectin-1 and Eps15 are simultaneously recruited to CEV inducing actin tails (AVI 1171 kb)
Supplementary Video 2
NPF motifs are required for viral recruitment of intersectin-1 (AVI 816 kb)
Supplementary Video 3
NPF motifs are required for viral recruitment of Eps15 (AVI 796 kb)
Supplementary Video 4
Loss of all three A36 NPF motifs results in significantly longer actin tails and a faster velocity of the virus (AVI 3238 kb)
Rights and permissions
About this article
Cite this article
Snetkov, X., Weisswange, I., Pfanzelter, J. et al. NPF motifs in the vaccinia virus protein A36 recruit intersectin-1 to promote Cdc42:N-WASP-mediated viral release from infected cells. Nat Microbiol 1, 16141 (2016). https://doi.org/10.1038/nmicrobiol.2016.141
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.141