Abstract
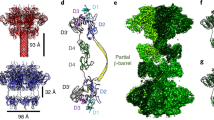
Clostridium difficile infection is the leading cause of hospital-acquired diarrhoea and pseudomembranous colitis. Disease is mediated by the actions of two toxins, TcdA and TcdB, which cause the diarrhoea, as well as inflammation and necrosis within the colon1,2. The toxins are large (308 and 270 kDa, respectively), homologous (47% amino acid identity) glucosyltransferases that target small GTPases within the host3,4. The multidomain toxins enter cells by receptor-mediated endocytosis and, upon exposure to the low pH of the endosome, insert into and deliver two enzymatic domains across the membrane. Eukaryotic inositol-hexakisphosphate (InsP6) binds an autoprocessing domain to activate a proteolysis event that releases the N-terminal glucosyltransferase domain into the cytosol. Here, we report the crystal structure of a 1,832-amino-acid fragment of TcdA (TcdA1832), which reveals a requirement for zinc in the mechanism of toxin autoprocessing and an extended delivery domain that serves as a scaffold for the hydrophobic α-helices involved in pH-dependent pore formation. A surface loop of the delivery domain whose sequence is strictly conserved among all large clostridial toxins is shown to be functionally important, and is highlighted for future efforts in the development of vaccines and novel therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lyerly, D. M., Krivan, H. C. & Wilkins, T. D. Clostridium difficile: its disease and toxins. Clin. Microbiol. Rev. 1, 1–18 (1988).
Kelly, C. P. & LaMont, J. T. Clostridium difficile—more difficult than ever. N. Engl. J. Med. 359, 1932–1940 (2008).
Voth, D. E. & Ballard, J. D. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18, 247–263 (2005).
Jank, T. & Aktories, K. Structure and mode of action of clostridial glucosylating toxins: the ABCD model. Trends Microbiol. 16, 222–229 (2008).
Lessa, F. C. et al. Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 825–834 (2015).
Lowy, I. et al. Treatment with monoclonal antibodies against Clostridium difficile toxins. N. Engl. J. Med. 362, 197–205 (2010).
Frey, S. M. & Wilkins, T. D. Localization of two epitopes recognized by monoclonal antibody PCG-4 on Clostridium difficile toxin A. Infect. Immun. 60, 2488–2492 (1992).
Orth, P. et al. Mechanism of action and epitopes of Clostridium difficile toxin B-neutralizing antibody bezlotoxumab revealed by X-ray crystallography. J. Biol. Chem. 289, 18008–18021 (2014).
Sauerborn, M., Leukel, P. & von Eichel-Streiber, C. The C-terminal ligand-binding domain of Clostridium difficile toxin A (TcdA) abrogates TcdA-specific binding to cells and prevents mouse lethality. FEMS Microbiol. Lett. 155, 45–54 (1997).
Frisch, C., Gerhard, R., Aktories, K., Hofmann, F. & Just, I. The complete receptor-binding domain of Clostridium difficile toxin A is required for endocytosis. Biochem. Biophys. Res. Commun. 300, 706–711 (2003).
Florin, I. & Thelestam, M. Lysosomal involvement in cellular intoxication with Clostridium difficile toxin B. Microb. Pathog. 1, 373–385 (1986).
Qa'Dan, M., Spyres, L. M. & Ballard, J. D. pH-induced conformational changes in Clostridium difficile toxin B. Infect. Immun. 68, 2470–2474 (2000).
Barth, H. et al. Low pH-induced formation of ion channels by Clostridium difficile toxin B in target cells. J. Biol. Chem. 276, 10670–10676 (2001).
Just, I. et al. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J. Biol. Chem. 270, 13932–13936 (1995).
Just, I. et al. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375, 500–503 (1995).
Egerer, M., Giesemann, T., Jank, T., Satchell, K. J. & Aktories, K. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J. Biol. Chem. 282, 25314–25321 (2007).
Reineke, J. et al. Autocatalytic cleavage of Clostridium difficile toxin B. Nature 446, 415–419 (2007).
Pruitt, R. N., Chambers, M. G., Ng, K. K., Ohi, M. D. & Lacy, D. B. Structural organization of the functional domains of Clostridium difficile toxins A and B. Proc. Natl Acad. Sci. USA 107, 13467–13472 (2010).
Albesa-Jove, D. et al. Four distinct structural domains in Clostridium difficile toxin B visualized using SAXS. J. Mol. Biol. 396, 1260–1270 (2010).
Ho, J. G., Greco, A., Rupnik, M. & Ng, K. K. Crystal structure of receptor-binding C-terminal repeats from Clostridium difficile toxin A. Proc. Natl Acad. Sci. USA 102, 18373–18378 (2005).
Murase, T. et al. Structural basis for antibody recognition in the receptor-binding domains of toxins A and B from Clostridium difficile. J. Biol. Chem. 289, 2331–2343 (2014).
Olling, A. et al. The combined repetitive oligopeptides of Clostridium difficile toxin A counteract premature cleavage of the glucosyl-transferase domain by stabilizing protein conformation. Toxins (Basel) 6, 2162–2176 (2014).
Zhang, Y., Hamza, T., Gao, S. & Feng, H. Masking autoprocessing of Clostridium difficile toxin A by the C-terminus combined repetitive oligo peptides. Biochem. Biophys. Res. Commun. 459, 259–263 (2015).
Jank, T., Giesemann, T. & Aktories, K. Clostridium difficile glucosyltransferase toxin B-essential amino acids for substrate binding. J. Biol. Chem. 282, 35222–35231 (2007).
Pruitt, R. N. et al. Structural determinants of Clostridium difficile toxin A glucosyltransferase activity. J. Biol. Chem. 287, 8013–8020 (2012).
Rupnik, M. et al. Characterization of the cleavage site and function of resulting cleavage fragments after limited proteolysis of Clostridium difficile toxin B (TcdB) by host cells. Microbiology 151, 199–208 (2005).
Kreimeyer, I. et al. Autoproteolytic cleavage mediates cytotoxicity of Clostridium difficile toxin A. Naunyn Schmiedebergs Arch. Pharmacol. 383, 253–262 (2011).
Pruitt, R. N. et al. Structure–function analysis of inositol hexakisphosphate-induced autoprocessing in Clostridium difficile toxin A. J. Biol. Chem. 284, 21934–21940 (2009).
Puri, A. W. et al. Rational design of inhibitors and activity-based probes targeting Clostridium difficile virulence factor TcdB. Chem. Biol. 17, 1201–1211 (2010).
Shen, A. et al. Defining an allosteric circuit in the cysteine protease domain of Clostridium difficile toxins. Nature Struct. Mol. Biol. 18, 364–371 (2011).
von Eichel-Streiber, C., Laufenberg-Feldmann, R., Sartingen, S., Schulze, J. & Sauerborn, M. Comparative sequence analysis of the Clostridium difficile toxins A and B. Mol. Gen. Genet. 233, 260–268 (1992).
Zhang, Z. et al. Translocation domain mutations affecting cellular toxicity identify the Clostridium difficile toxin B pore. Proc. Natl Acad. Sci. USA 111, 3721–3726 (2014).
Genisyuerek, S. et al. Structural determinants for membrane insertion, pore formation and translocation of Clostridium difficile toxin B. Mol. Microbiol. 79, 1643–1654 (2011).
Choe, S. et al. The crystal structure of diphtheria toxin. Nature 357, 216–222 (1992).
Wang, J. & London, E. The membrane topography of the diphtheria toxin T domain linked to the a chain reveals a transient transmembrane hairpin and potential translocation mechanisms. Biochemistry 48, 10446–10456 (2009).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Teichert, M., Tatge, H., Schoentaube, J., Just, I. & Gerhard, R. Application of mutated Clostridium difficile toxin A for determination of glucosyltransferase-dependent effects. Infect. Immun. 74, 6006–6010 (2006).
Chumbler, N. M. et al. Clostridium difficile toxin B causes epithelial cell necrosis through an autoprocessing-independent mechanism. PLoS Pathogens 8, e1003072 (2012).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Otwinowski, Z. & Minor, W. in Methods in Enzymology Vol. 276: Macromolecular Crystallography, part A (eds Carter, C. W. Jr & Sweets, R. M.) 307–326 (Academic, 1997).
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D 60, 2126–2132 (2004).
Ravel, B. & Newville, M. ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J. Synchrotron Radiat. 12, 537–541 (2005).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675 (2012).
Acknowledgements
This research was supported by NIAID of the National Institutes of Health (award no. R01AI095755 to D.B.L.) and NIGMS (award no. R01GM042569 to D.P.G.). The authors thank staff at the LS-CAT beamline for help with data collection. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under contract no. DE-AC02-06CH11357. Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant no. 085P1000817). Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the US DOE, Office of Science, Office of Basic Energy Sciences (contract no. DE-AC02-98CH10886). Operations at the NSLS beamline X3B were supported by NIH P30-EB009998.
Author information
Authors and Affiliations
Contributions
S.A.R. crystallized TcdA1832. S.A.R., B.W.S. and D.B.L. determined the TcdA1832 structure. S.A.R., N.M.C. and M.A.F. generated expression clones and purified proteins. N.M.C. and M.A.F. conducted autoprocessing, Rac1 glucosylation and cell binding assays. Z.Z. conducted viability and Rb+ release assays. N.M.C. prepared samples for ICP-MS assays and J.P.L. performed the assays. E.F. performed XAS measurements. All authors were involved in data analysis and assisted in editing the manuscript. D.B.L. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Tables 1–4, Figures 1–5 and References (PDF 1613 kb)
Supplementary Video 1
A cartoon representation of the TcdA1832 crystal structure coloured as in Figure 1 and rotating about the vertical axis. (MOV 9685 kb)
Supplementary Video 2
A hypothetical trajectory of movement between the apo- and InsP6-bound structures of the TcdA APD highlights significant structural changes in the InsP6 binding site, the beta-flap, and the APD active site. The trajectory for the protein (coloured as in Figure 2) was calculated in Chimera (ref. 36). (MOV 4776 kb)
Rights and permissions
About this article
Cite this article
Chumbler, N., Rutherford, S., Zhang, Z. et al. Crystal structure of Clostridium difficile toxin A. Nat Microbiol 1, 15002 (2016). https://doi.org/10.1038/nmicrobiol.2015.2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2015.2
This article is cited by
-
Unconventional structure and mechanisms for membrane interaction and translocation of the NF-κB-targeting toxin AIP56
Nature Communications (2023)
-
Identification of TFPI as a receptor reveals recombination-driven receptor switching in Clostridioides difficile toxin B variants
Nature Communications (2022)
-
Clostridioides difficile toxins: mechanisms of action and antitoxin therapeutics
Nature Reviews Microbiology (2022)
-
Gastrobodies are engineered antibody mimetics resilient to pepsin and hydrochloric acid
Communications Biology (2021)
-
Secondary-structure switch regulates the substrate binding of a YopJ family acetyltransferase
Nature Communications (2021)