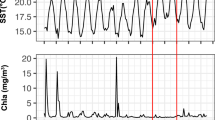
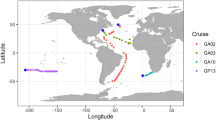
Abstract
Marine viruses are critical drivers of ocean biogeochemistry, and their abundances vary spatiotemporally in the global oceans, with upper estimates exceeding 108 per ml. Over many years, a consensus has emerged that virus abundances are typically tenfold higher than microbial cell abundances. However, the true explanatory power of a linear relationship and its robustness across diverse ocean environments is unclear. Here, we compile 5,508 microbial cell and virus abundance estimates from 22 distinct marine surveys and find substantial variation in the virus-to-microbial cell ratio, in which a 10:1 model has either limited or no explanatory power. Instead, virus abundances are better described as nonlinear, power-law functions of microbial cell abundances. The fitted scaling exponents are typically less than 1, implying that the virus-to-microbial cell ratio decreases with microbial cell density, rather than remaining fixed. The observed scaling also implies that viral effect sizes derived from ‘representative’ abundances require substantial refinement to be extrapolated to regional or global scales.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
03 October 2017
The original publication of this Article included analysis of virus and microbial cell abundances and virus-to-microbial cell ratios. Data in the Article came from 25 studies intended to be exclusively from marine sites. However, 3 of the studies included in the original unified dataset were erroneously classified as marine sites during compilation. The records with mis-recorded longitude and latitude values were, in fact, taken from inland, freshwater sources. The three inland, freshwater datasets are ELA, TROUT and SWAT. The data from these three studies represent 163 of the 5,671 records in the original publication. In the updated version of the Article, all analyses have been recalculated using the same statistical analysis pipeline released via GitHub as part of the original publication. Removal of the three studies reduces the unified dataset to 5,508 records. Analyses involving all grouped datasets have been updated with changes noted in each figure. All key results remain qualitatively unchanged. All data and scripts used in this correction have been made available as a new, updated GitHub release to reflect the updated dataset and figures.
References
Thingstad, T. F. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45, 1320–1328 (2000).
Suttle, C. A. Viruses in the sea. Nature 437, 356–361 (2005).
Brussaard, C. P. D. et al. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 20, 575–578 (2008).
Rohwer, F. & Thurber, R. V. Viruses manipulate the marine environment. Nature 459, 207–212 (2009).
Weitz, J. S. & Wilhelm, S. W. Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol. Reports 4, 17 (2012).
Jover, L. F., Effler, T. C., Buchan, A., Wilhelm, S. W. & Weitz, J. S. The elemental composition of virus particles: implications for marine biogeochemical cycles. Nature Rev. Microbiol. 12, 519–528 (2014).
Weitz, J. S., Hartman, H. & Levin, S. A. Coevolutionary arms races between bacteria and bacteriophage. Proc. Natl Acad. Sci. USA 102, 9535–9540 (2005).
Avrani, S., Schwartz, D. A. & Lindell, D. Virus-host swinging party in the oceans: incorporating biological complexity into paradigms of antagonistic coexistence. Mobile Genet. Elements 2, 88–95 (2012).
Payet, J. P. & Suttle, C. A. To kill or not to kill: the balance between lytic and lysogenic viral infection is driven by trophic status. Limnol. Oceanogr. 58, 465–474 (2013).
Murray, A. G. & Jackson, G. A. Viral dynamics: a model of the effects of size, shape, motion and abundance of single-celled planktonic organisms and other particles. Marine Ecol. Progr. Ser. 89, 103–116 (1992).
Bergh, O., Borhseim, K. Y., Bratbak, G. & Heldal, M. High abundance of viruses found in aquatic environments. Nature 340, 467–468 (1989).
Maranger, R. & Bird, D. F. Viral abundance in aquatic systems—a comparison between marine and fresh-waters. Marine Ecol. Progr. Ser. 1210, 217–226 (1995).
Wommack, K. E. & Colwell, R. R. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64, 69–114 (2000).
Weinbauer, M. G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181 (2004).
Mojica, K. D. A. et al. Phytoplankton community structure in relation to vertical stratification along a north–south gradient in the Northeast Atlantic Ocean. Limnol. Oceanogr. 60, 1498–1521 (2015).
Edwards, R. A. & Rohwer, F. Viral metagenomics. Nature Rev. Microbiol. 3, 504–510 (2005).
Brum, J. R., Hurwitz, B. L., Schofield, O., Ducklow, H. W. & Sullivan, M. B. Seasonal time bombs: dominant temperate viruses affect southern ocean microbial dynamics. ISME J. http://dx.doi.org/10.1038/ismej.2015.125 (2015).
Weitz, J. S. et al. Phage–bacteria infection networks. Trends Microbiol. 21, 82–91 (2013).
Danovaro, R. et al. Marine viruses and global climate change. FEMS Microbiol. Rev. 35, 993–1034 (2011).
Morel, A. & Berthon, J. Surface pigments, algal biomass profiles, and potential production of the euphotic layer: relationships reinvestigated in view of remote-sensing applications. Limnol. Oceanogr. 340, 1545–1562 (1989).
Parsons, R. J., Breitbart, M., Lomas, M. W. & Carlson, C. A. Ocean time-series reveals recurring seasonal patterns of virioplankton dynamics in the northwestern Sargasso Sea. ISME J. 6, 273–284 (2011).
Fuhrman, J. A. et al. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl Acad. Sci. USA 103, 13104–13109 (2006).
De Corte, D., Sintes, E., Yokokawa, T., Reinthaler, T. & Herndl, G. J. Links between viruses and prokaryotes throughout the water column along a north Atlantic latitudinal transect. ISME J. 60, 1566–1577 (2012).
Li, W. K. W. & Dickie, P. M. Monitoring phytoplankton, bacterioplankton, and virioplankton in a coastal inlet (Bedford Basin) by flow cytometry. Cytometry 440, 236–246 (2001).
Yang, Y., Yokokawa, T., Motegi, C. & Nagata, T. Large-scale distribution of viruses in deep waters of the pacific and southern oceans. Aquatic Microbial Ecol. 710, 193–202 (2013).
Clasen, J. L., Brigden, S. M., Payet, J. P. & Suttle, C. A. Evidence that viral abundance across oceans and lakes is driven by different biological factors. Freshwater Biol. 53, 1090–1100 (2008).
Balsom, A. L. Macroinfaunal Community Composition and Biomass, and Bacterial and Viral Abundances from the Gulf of Alaska to the Canadian Archipelago: a Biodiversity Study. MSc thesis, Univ. Tennessee (2003).
Strzepek, R. F. et al. Spinning the ‘ferrous wheel’: the importance of the microbial community in an iron budget during the FeCycle experiment. Global Biogeochem. Cycles 19, GB4S26 (2005).
Matteson, A. R. et al. Production of viruses during a spring phytoplankton bloom in the South Pacific Ocean near New Zealand. FEMS Microbiol. Ecol. 79, 709–719 (2012).
Rowe, J. M. et al. Constraints on viral production in the Sargasso Sea and North Atlantic. Aquatic Microbial Ecol. 520, 233–244 (2008).
Wilhelm, S. W. et al. UV radiation induced DNA damage in marine viruses along a latitudinal gradient in the southeastern Pacific Ocean. Aquatic Microbial Ecol. 310, 1–8 (2003).
Wang, K., Eric Wommack, K. & Chen, F. Abundance and distribution of Synechococcus spp. and cyanophages in the Chesapeake Bay. Appl. Environ. Microbiol. 770, 7459–7468 (2011).
Suttle, C. A. Marine viruses—major players in the global ecosystem. Nature Rev. Microbiol. 5, 801–812 (2007).
Danovaro, R. et al. Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454, 1084–1U27 (2008).
Brum, J. R. & Sullivan, M. B. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nature Rev. Microbiol. 130, 147–159 (2015).
Weitz, J. S. et al. A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J. 90, 1352–1364 (2015).
Suttle, C. A. & Chan, A. M. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Marine Ecol. Progr. Ser. 92, 99–109 (1993).
De Paepe, M. & Taddei, F. Viruses’ life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 4, e193 (2006).
Thingstad, T. F. & Lignell, R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquatic Microbial Ecol. 13, 19–27 (1997).
Giovannoni, S., Tempterton, B. & Zhao, Y. Nature 499, E4–E5 (2013).
Carreira, C., Larsen, M., Glud, R. N., Brussaard, C. P. D. & Middelboe, M. Heterogeneous distribution of prokaryotes and viruses at the microscale in a tidal sediment. Aquatic Microbial Ecol. 69, 183–192 (2013).
Bratbak, G. & Heldal, M. in Molecular Ecology of Aquatic Microbes Vol. 238 (ed. Joint, I. ) 249–264 (Springer-Verlag, 1995).
Williamson, S. J., Houchin, L. A., McDaniel, L. & Paul, J. H. Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl. Environ. Microbiol. 68, 4307–4314 (2002).
Hatton, I. A. et al. The predator–prey power law: biomass scaling across terrestrial and aquatic biomes. Science 349, aac6284 (2015).
Acknowledgements
This work was supported by National Science Foundation (NSF) grants OCE-1233760 (to J.S.W.) and OCE-1061352 (to A.B. and S.W.W.), a Career Award at the Scientific Interface from the Burroughs Wellcome Fund (to J.S.W.) and a Simons Foundation SCOPE grant (to J.S.W.). This work was conducted as part of the Ocean Viral Dynamics Working Group at the National Institute for Mathematical and Biological Synthesis, sponsored by the National Science Foundation through NSF Award DBI-1300426, with additional support from The University of Tennessee, Knoxville.
Author information
Authors and Affiliations
Contributions
C.H.W. developed the code and implemented all scripts, analysed data, performed statistical analysis and contributed to writing the manuscript. D.S. developed and reviewed the code, analysed data, performed statistical analysis and provided feedback on the manuscript. A.B., J.F., J.T.L., M.M., C.A.S., C.S., W.H.W. and K.E.W. contributed to the design and implementation of the study, the assessment and collection of data sets, and provided feedback on the manuscript. C.P.D.B. and J.F.F. contributed to the assessment and collection of data sets and provided feedback on the manuscript. S.W.W. co-led the design of the study, led the data collection and assessment component, and contributed to writing the manuscript. J.S.W. co-led the design of the study, led the code and statistical analysis component, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Containing Supplementary Text, Tables 1–6, Figures 1–6 and References (PDF 1140 kb)
Supplementary Data 1
Supplementary Dataset (XLS 646 kb)
Rights and permissions
About this article
Cite this article
Wigington, C., Sonderegger, D., Brussaard, C. et al. Re-examination of the relationship between marine virus and microbial cell abundances. Nat Microbiol 1, 15024 (2016). https://doi.org/10.1038/nmicrobiol.2015.24
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2015.24
This article is cited by
-
Benchmarking bioinformatic virus identification tools using real-world metagenomic data across biomes
Genome Biology (2024)
-
Counting dots or counting reads? Complementary approaches to estimate virus-to-microbe ratios
The ISME Journal (2023)
-
Geochemical constraints on bacteriophage infectivity in terrestrial environments
ISME Communications (2023)
-
Metagenome-derived virus-microbe ratios across ecosystems
The ISME Journal (2023)
-
Virus-to-prokaryote ratio in spring waters along a gradient of natural radioactivity
Hydrobiologia (2023)