Abstract
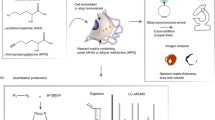
We present an extracellular matrix (ECM) microarray platform for the culture of patterned cells atop combinatorial matrix mixtures. This platform enables the study of differentiation in response to a multitude of microenvironments in parallel. The fabrication process required only access to a standard robotic DNA spotter, off-the-shelf materials and 1,000 times less protein than conventional means of investigating cell-ECM interactions. To demonstrate its utility, we applied this platform to study the effects of 32 different combinations of five extracellular matrix molecules (collagen I, collagen III, collagen IV, laminin and fibronectin) on cellular differentiation in two contexts: maintenance of primary rat hepatocyte phenotype indicated by intracellular albumin staining and differentiation of mouse embryonic stem (ES) cells toward an early hepatic fate, indicated by expression of a β-galactosidase reporter fused to the fetal liver-specific gene, Ankrd17 (also known as gtar). Using this technique, we identified combinations of ECM that synergistically impacted both hepatocyte function and ES cell differentiation. This versatile technique can be easily adapted to other applications, as it is amenable to studying almost any insoluble microenvironmental cue in a combinatorial fashion and is compatible with several cell types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ding, S. et al. Synthetic small molecules that control stem cell fate. Proc. Natl. Acad. Sci. USA 100, 7632–7637 (2003).
Revzin, A. et al. Designing a hepatocellular microenvironment with protein microarraying and poly(ethylene glycol) photolithography. Langmuir 20, 2999–3005 (2004).
Prudhomme, W., Daley, G.Q., Zandstra, P. & Lauffenburger, D.A. Multivariate proteomic analysis of murine embryonic stem cell self-renewal versus differentiation signaling. Proc. Natl. Acad. Sci. USA 101, 2900–2905 (2004).
Anderson, D.G., Levenberg, S. & Langer, R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat. Biotechnol. 22, 863–866 (2004).
Meredith, J.E. Jr. et al. The regulation of growth and intracellular signaling by integrins. Endocr. Rev. 17, 207–220 (1996).
Comoglio, P.M., Boccaccio, C. & Trusolino, L. Interactions between growth factor receptors and adhesion molecules: breaking the rules. Curr. Opin. Cell Biol. 15, 565–571 (2003).
Hall, H.G., Farson, D.A. & Bissell, M.J. Lumen formation by epithelial cell lines in response to collagen overlay: a morphogenetic model in culture. Proc. Natl. Acad. Sci. USA 79, 4672–4676 (1982).
Dunn, J.C., Yarmush, M.L., Koebe, H.G. & Tompkins, R.G. Hepatocyte function and extracellular matrix geometry: long-term culture in a sandwich configuration. FASEB J. 3, 174–177 (1989).
Cukierman, E., Pankov, R., Stevens, D.R. & Yamada, K.M. Taking cell-matrix adhesions to the third dimension. Science 294, 1708–1712 (2001).
Baatout, S. Endothelial differentiation using Matrigel (review). Anticancer Res. 17, 451–455 (1997).
Lindberg, K. & Badylak, S.F. Porcine small intestinal submucosa (SIS): a bioscaffold supporting in vitro primary human epidermal cell differentiation and synthesis of basement membrane proteins. Burns 27, 254–266 (2001).
Lin, P., Chan, W.C.W., Badylak, S. & Bhatia, S. Assessing the function of porcine liver-derived matrix: applications to hepatic tissue engineering. Tissue Eng. 10, 1046–1053 (2004).
Mann, B.K., Gobin, A.S., Tsai, A.T., Schmedlen, R.H. & West, J.L. Smooth muscle cell growth in photopolymerized hydrogels with cell adhesive and proteolytically degradable domains: synthetic ECM analogs for tissue engineering. Biomaterials 22, 3045–3051 (2001).
Hern, D.L. & Hubbell, J.A. Incorporation of adhesion peptides into nonadhesive hydrogels useful for tissue resurfacing. J. Biomed. Mater. Res. 39, 266–276 (1998).
Anseth, K.S., Shastri, V.R. & Langer, R. Photopolymerizable degradable polyanhydrides with osteocompatibility. Nat. Biotechnol. 17, 156–159 (1999).
Singhvi, R. et al. Engineering cell-shape and function. Science 264, 696–698 (1994).
Chen, C.S., Mrksich, M., Huang, S., Whitesides, G.M. & Ingber, D.E. Geometric control of cell life and death. Science 276, 1425–1428 (1997).
Bhatia, S.N., Yarmush, M.L. & Toner, M. Controlling cell interactions by micropatterning in co-cultures: hepatocytes and 3T3 fibroblasts. J. Biomed. Mater. Res. 34, 189–199 (1997).
Kane, R.S., Takayama, S., Ostuni, E., Ingber, D.E. & Whitesides, G.M. Patterning proteins and cells using soft lithography. Biomaterials 20, 2363–2376 (1999).
MacBeath, G. & Schreiber, S.L. Printing proteins as microarrays for high-throughput function determination. Science 289, 1760–1763 (2000).
Houseman, B.T. & Mrksich, M. Carbohydrate arrays for the evaluation of protein binding and enzymatic modification. Chem. Biol. 9, 443–454 (2002).
Falsey, J.R., Renil, M., Park, S., Li, S.J. & Lam, K.S. Peptide and small molecule microarray for high throughput cell adhesion and functional assays. Bioconj. Chem. 12, 346–353 (2001).
Ziauddin, J. & Sabatini, D.M. Microarrays of cells expressing defined cDNAs. Nature 411, 107–110 (2001).
Mousses, S. et al. RNAi microarray analysis in cultured mammalian cells. Genome Res. 13, 2341–2347 (2003).
Forrester, L.M. et al. An induction gene trap screen in embryonic stem cells: Identification of genes that respond to retinoic acid in vitro. Proc. Natl. Acad. Sci. USA 93, 1677–1682 (1996).
Watt, A.J. et al. A gene trap integration provides an early in situ marker for hepatic specification of the foregut endoderm. Mech. Dev. 100, 205–215 (2001).
Sudhakaran, P.R., Stamatoglou, S.C. & Hughes, R.C. Modulation of protein synthesis and secretion by substratum in primary cultures of rat hepatocytes. Exp. Cell Res. 167, 505–516 (1986).
Hughes, R.C. & Stamatoglou, S.C. Adhesive interactions and the metabolic activity of hepatocytes. J. Cell Sci. Suppl. 8, 273–291 (1987).
Freire, E. & Coelho-Sampaio, T. Self-assembly of laminin induced by acidic pH. J. Biol. Chem. 275, 817–822 (2000).
Nelson, C.M., Raghavan, S., Tan, J.L. & Chen, C.S. Degradation of micropatterned surfaces by cell-dependent and -independent processes. Langmuir 19, 1493–1499 (2003).
Angenendt, P., Glokler, J., Murphy, D., Lehrach, H. & Cahill, D.J. Toward optimized antibody microarrays: a comparison of current microarray support materials. Anal. Biochem. 309, 253–260 (2002).
Revzin, A. et al. Fabrication of poly(ethylene glycol) hydrogel microstructures using photolithography. Langmuir 17, 5440–5447 (2001).
Koh, W.G., Revzin, A. & Pishko, M.V. Poly(ethylene glycol) hydrogel microstructures encapsulating living cells. Langmuir 18, 2459–2462 (2002).
Kim, C.O., Hong, S.-Y., Kim, M., Park, S.-M. & Park, J.W. Modification of indium-tin oxide (ITO) glass with aziridine provides a surface of high amine density. J. Colloid Interface Sci. 277, 499–504 (2004).
Timofeev, E., Kochetkova, S.V., Mirzabekov, A.D. & Florentiev, V.L. Regioselective immobilization of short oligonucleotides to acrylic copolymer gels. Nucleic Acids Res. 24, 3142–3148 (1996).
Massia, S.P. & Hubbell, J.A. An RGD spacing of 440 nm is sufficient for integrin α V β 3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation. J. Cell Biol. 114, 1089–1100 (1991).
Mooney, D. et al. Switching from differentiation to growth in hepatocytes: control by extracellular matrix. J. Cell. Physiol. 151, 497–505 (1992).
Pinkse, G.G. et al. Hepatocyte survival depends on β1-integrin-mediated attachment of hepatocytes to hepatic extracellular matrix. Liver Int. 24, 218–226 (2004).
Martinez-Hernandez, A. & Amenta, P.S. The hepatic extracellular matrix. I. Components and distribution in normal liver. Virchows Arch. A Pathol. Anat. Histopathol. 423, 1–11 (1993).
Stamatoglou, S.C., Enrich, C., Manson, M.M. & Hughes, R.C. Temporal changes in the expression and distribution of adhesion molecules during liver development and regeneration. J. Cell Biol. 116, 1507–1515 (1992).
Liu Tsang, V. Three-dimensional photopatterning of hydrogels containing living cells for hepatic tissue engineering. Ph.D. dissertation, University of California San Diego, La Jolla (2004).
Werb, Z., Tremble, P.M., Behrendtsen, O., Crowley, E. & Damsky, C.H. Signal transduction through the fibronectin receptor induces collagenase and stromelysin gene expression. J. Cell Biol. 109, 877–889 (1989).
Pacifici, R. et al. Ligand binding to monocyte α5β1 integrin activates the α2β1 receptor via the α5 subunit cytoplasmic domain and protein kinase C. J. Immunol. 153, 2222–2233 (1994).
Diegelmann, R.F., Guzelian, P.S., Gay, R. & Gay, S. Collagen formation by the hepatocyte in primary monolayer culture and in vivo. Science 219, 1343–1345 (1983).
Fashena, S.J. & Thomas, S.M. Signalling by adhesion receptors. Nat. Cell Biol. 2, E225–E229 (2000).
Schwartz, M.A. & Ginsberg, M.H. Networks and crosstalk: integrin signalling spreads. Nat. Cell Biol. 4, E65–E68 (2002).
Pacifici, R. et al. Collagen-induced release of interleukin 1 from human blood mononuclear cells. Potentiation by fibronectin binding to the α5β1 integrin. J. Clin. Invest. 89, 61–67 (1992).
Kim, S., Harris, M. & Varner, J.A. Regulation of integrin αvβ3-mediated endothelial cell migration and angiogenesis by integrin α5β1 and protein kinase A. J. Biol. Chem. 275, 33920–33928 (2000).
Rathjen, J. et al. Directed differentiation of pluripotent cells to neural lineages: homogeneous formation and differentiation of a neurectoderm population. Development 129, 2649–2661 (2002).
Ying, Q.L., Stavridis, M., Griffiths, D., Li, M. & Smith, A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186 (2003).
Ivanova, N.B. et al. A stem cell molecular signature. Science 298, 601–604 (2002).
Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R.C. & Melton, D.A. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298, 597–600 (2002).
Kelly, D.L. & Rizzino, A. DNA microarray analyses of genes regulated during the differentiation of embryonic stem cells. Mol. Reprod. Dev. 56, 113–123 (2000).
Hamazaki, T. et al. Hepatic maturation in differentiating embryonic stem cells in vitro. FEBS Lett. 497, 15–19 (2001).
Yamada, T. et al. In vitro differentiation of embryonic stem cells into hepatocyte-like cells identified by cellular uptake of indocyanine green. Stem Cells 20, 146–154 (2002).
Chinzei, R. et al. Embryoid-body cells derived from a mouse embryonic stem cell line show differentiation into functional hepatocytes. Hepatology 36, 22–29 (2002).
Ishizaka, S. et al. Development of hepatocytes from ES cells after transfection with the HNF-3β gene. FASEB J. 16, 1444–1446 (2002).
Jones, E.A., Tosh, D., Wilson, D.I., Lindsay, S. & Forrester, L.M. Hepatic differentiation of murine embryonic stem cells. Exp. Cell Res. 272, 15–22 (2002).
Subramanian, S. & Srienc, F. Quantitative analysis of transient gene expression in mammalian cells using the green fluorescent protein. J. Biotechnol. 49, 137–151 (1996).
Arenkov, P. et al. Protein microchips: use for immunoassay and enzymatic reactions. Anal. Biochem. 278, 123–131 (2000).
Acknowledgements
We would like to thank L. Forrester (University of Edinburgh) for providing the I114 cell line, C. Largent (Cel Associates) and D. Schabacker (Argonne) for useful discussions about acrylamide-gel pad fabrication, T. Martinsky (Telechem) for printing parameters, J. Norwich (S. Chien Lab), R. Agustin (imaging, J. Price Lab), J. Emond and R. Lieber (biostatistics), S. Khetani (cell patterning), J. Felix (hepatocyte isolation) and O. Tran. Funding was generously provided by the US National Institutes of Health, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Science Foundation's Faculty Early Career Development (CAREER) Program and the David and Lucile Packard Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Spotted ECM mediates cell attachment (PDF 227 kb)
Rights and permissions
About this article
Cite this article
Flaim, C., Chien, S. & Bhatia, S. An extracellular matrix microarray for probing cellular differentiation. Nat Methods 2, 119–125 (2005). https://doi.org/10.1038/nmeth736
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth736
This article is cited by
-
Morphogenesis beyond in vivo
Nature Reviews Physics (2023)
-
Microneedle array facilitates hepatic sinusoid construction in a large-scale liver-acinus-chip microsystem
Microsystems & Nanoengineering (2023)
-
Reconstructing the pulmonary niche with stem cells: a lung story
Stem Cell Research & Therapy (2022)
-
A guide to the organ-on-a-chip
Nature Reviews Methods Primers (2022)
-
Delineating cooperative effects of Notch and biomechanical signals on patterned liver differentiation
Communications Biology (2022)