Nat. Methods 7, 237–242 (2010); published online 14 Feburary 2010; addendum published after print 29 September 2010.
After the publication of our paper, we identified a mistake in Table 1 regarding the comparison of our program, Waltz, to the program 3D profile1 (ref. 25 in our paper); we cited the wrong name and reference of the algorithm in the right column. This error has been corrected after print to refer to the algorithm we actually used, the method described in reference 2 (ref. 40 in the corrected paper). However, as the 3D profile1 method developed in the Eisenberg laboratory has a long-standing good reputation as an amyloid prediction tool, here we compare it to Waltz. An improved version of 3D profile3 was published about a week and a half before our paper, so for complete transparency we also compare Waltz to the improved 3D profile algorithm.
In Table 1 we list all predicted peptides and scores or energies, respectively, comparing Waltz (threshold 77, running on our webserver at http://waltz.switchlab.org/) with the 3D profile1 scores at the ZipperDB website (http://services.mbi.ucla.edu/zipperdb; energy threshold was −23; additional shape complementarity > 0.7 for the 3D profile 2010 version3). The sensitivity of 3D profile on our sup35 positive set was 67% (75% if one includes prediction of a hexapeptide that is almost but not fully included in the tested decapeptide).
However, the higher sensitivity of 3D profile comes at a cost of lower specificity (more false positives). To estimate the rate of false positives, we derived a reliable negative set from our experimental data for sup35, which included all decapeptides that did not form fibers under the unified experimental conditions and did not overlap with any positively tested one (31 in total). However, we cannot draw hard conclusions as the availability of bona fide experimental data is typically limiting and these numbers are too low for a good general comparison. An additional complication is that 3D profile is designed to predict hexapeptides; as next best approximation we defined the best score or energy of a fully included hexapeptide as prediction for the respective peptides. Owing to this limitation and the fact that well-predicted hexapeptides may actually form amyloid fibers and the longer decapeptide does not, it may be wiser to exclude such peptides in an alternative comparison with only 26 'negative' peptides, the reduced benchmark set ('–5') (Table 1).
Sensitivities of predictors should either be compared at similar levels of specificity (as should be done in consensus methods, such as AmylPred4), or one needs to consider both sensitivity and specificity together. Established measures for this are the Matthew correlation coefficient and the probability excess5. Probability excess has the additional advantage that it is also independent of set size inequalities6, which are not considered in other measures such as accuracy and precision.
The resulting performance statistics are reported in Table 2. Although 3D profile 2006 version1 predicted several additional false positives compared to Waltz, the improved 3D profile 2010 version3 filtered out several of these. Considering the possibility that high-scoring hexapeptides may indeed form fibers outside of the experimentally tested decapeptide context, the performances of Waltz and 3D profile (2010 version)3 become comparable over the reduced benchmark set ('–5'). In fact, the observed differences may well be within the error of performance estimation given the small benchmark set.
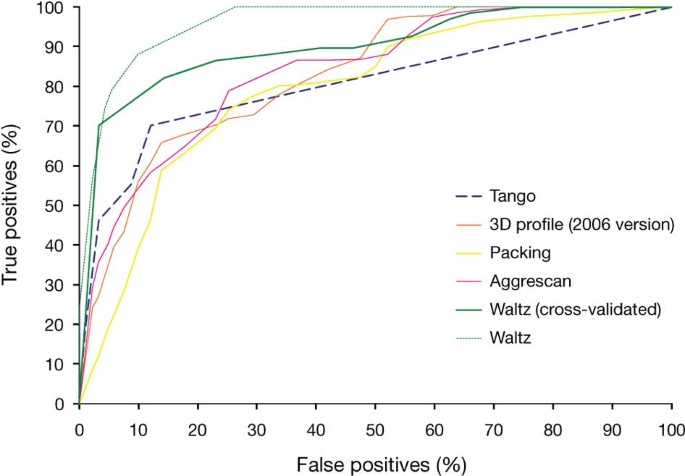
We also performed a receiver operating characteristics (ROC) curve analysis to benchmark the performance of Waltz, 3D profile 2006 version1, Tango7, Packing8 and Aggrescan9 on the AmylHex dataset1 (Fig. 1). The AmylHex dataset is an experimentally validated set of hexapeptides containing 67 positive (amyloid forming) and 91 negative (non–fiber forming) examples. Although 3D profile and the other methods in this benchmark were not subjected to cross-validation, we additionally scrutinized Waltz using rigorous cross-validation criteria as outlined in Supplementary Notes 3 and 4 of our original paper. We emphasize that the 3D profile method1 in this ROC curve was the version from 2006; we did not test the performance of the improved 3D profile3 method.
Our and others' recent work has additionally contributed several new experimentally verified examples, which should form the basis of an enlarged benchmark set to allow standardized ROC comparison of amyloid predictors by all interested groups in the future.
References
Thompson, M.J. et al. The 3D profile method for identifying fibril-forming segments of proteins. Proc. Natl. Acad. Sci. USA 103, 4074–4078 (2006).
Zhang, Z.Q., Chen, H. & Lai, L.H. Identification of amyloid fibril-forming segments based on structure and residue-based statistical potential. Bioinformatics 23, 2218–2225 (2007).
Goldschmidt, L., Teng, P.K., Riek, R. & Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA 107, 3487–3492 (2010).
Hamodrakas, S.J., Liappa, C. & Iconomidou, V.A. Consensus prediction of amyloidogenic determinants in amyloid fibril-forming proteins. Int. J. Biol. Macromol. 41, 295–300 (2007).
Sirota, F.L. et al. Parameterization of disorder predictors for large-scale applications requiring high specificity by using an extended benchmark dataset. BMC Genomics 11, (Suppl. 1) S15 (2010).
Yang, Z.R., Thomson, R., McNeil, P. & Esnouf, R.M. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21, 3369–3376 (2005).
Fernandez-Escamilla, A.M., Rousseau, F., Schymkowitz, J. & Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22, 1302–1306 (2004).
Galzitskaya, O.V., Garbuzynskiy, S.O. & Lobanov, M.Y. Prediction of amyloidogenic and disordered regions in protein chains. PLOS Comput. Biol. 2, e177 (2006).
Conchillo-Solé, O. et al. AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinformatics 8, 65 (2007).
Rights and permissions
About this article
Cite this article
Maurer-Stroh, S., Debulpaep, M., Kuemmerer, N. et al. Addendum: Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods 7, 855–857 (2010). https://doi.org/10.1038/nmeth1010-855b
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth1010-855b